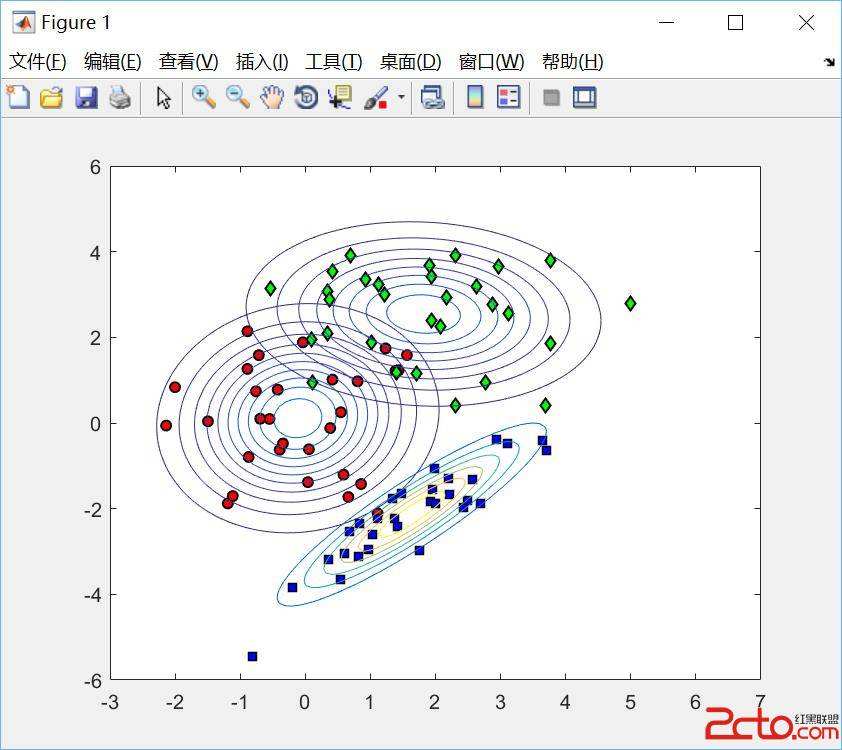
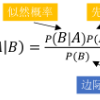
Whether based on models, training data or a combination, classifiers place (possibly complex) input data into one of a relatively small number of output categories. In this paper, we study the structure of the boundary--those points for which a neighbor is classified differently--in the context of an input space that is a graph, so that there is a concept of neighboring inputs, The scientific setting is a model-based naive Bayes classifier for DNA reads produced by Next Generation Sequencers. We show that the boundary is both large and complicated in structure. We create a new measure of uncertainty, called Neighbor Similarity, that compares the result for a point to the distribution of results for its neighbors. This measure not only tracks two inherent uncertainty measures for the Bayes classifier, but also can be implemented, at a computational cost, for classifiers without inherent measures of uncertainty.
翻译:无论是基于模型、 培训数据还是组合, 分类者将( 可能是复杂的) 输入数据输入到数量相对较少的产出类别之一中。 在本文中, 我们研究了相邻者被不同分类的边界点的结构, 以图形输入空间为背景, 这样就有了相邻投入的概念, 科学环境是一个基于模型的天真贝斯分类器, DNA是由下一代序列器编写的。 我们显示边界在结构上既大又复杂。 我们创造了一种新的不确定性度量, 称为邻居相似性, 将结果点与邻居结果分布进行比较 。 这一度量不仅跟踪了贝斯分类者的两个固有的不确定性度, 而且可以计算成本, 用于没有内在不确定性度的分类者 。