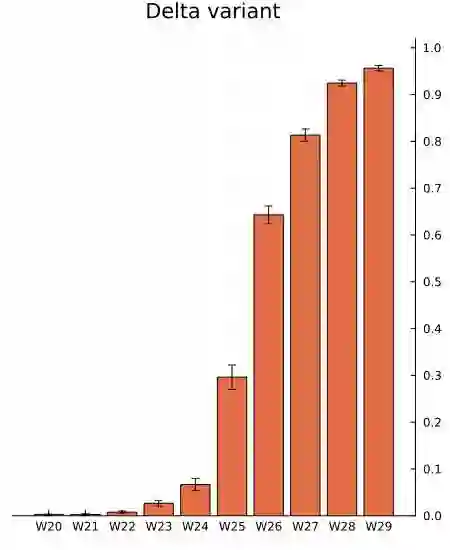
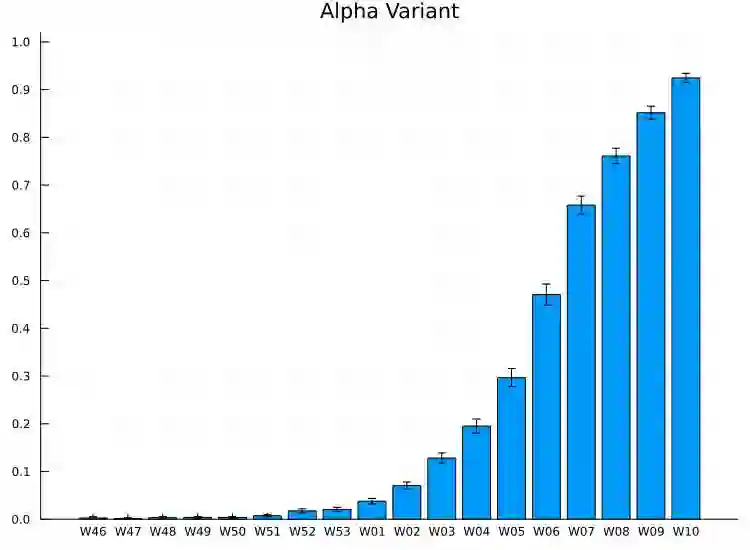
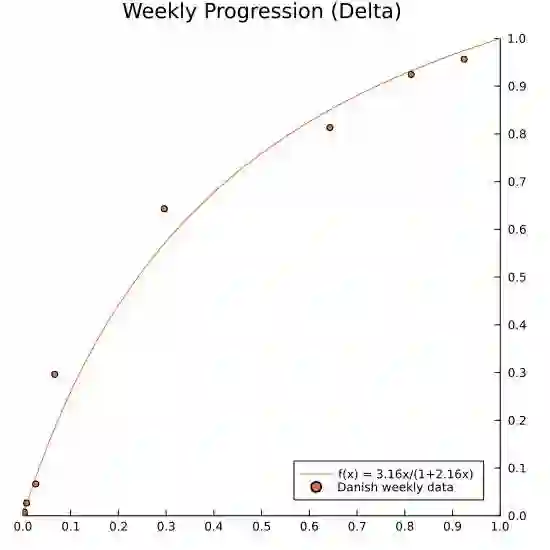
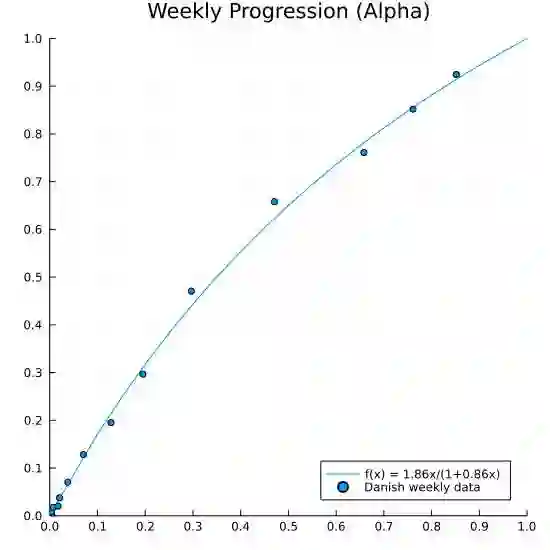
We propose a simple dynamic model for estimating the relative contagiousness of two virus variants. Maximum likelihood estimation and inference is conveniently invariant to variation in the total number of cases over the sample period and can be expressed as a logistic regression. We apply the model to Danish SARS-CoV-2 variant data. We estimate the reproduction numbers of Alpha and Delta to be larger than that of the ancestral variant by a factor of 1.51 [CI 95%: 1.50, 1.53] and 3.28 [CI 95%: 3.01, 3.58], respectively. In a predominately vaccinated population, we estimate Omicron to be 3.15 [CI 95%: 2.83, 3.50] times more infectious than Delta. Forecasting the proportion of an emerging virus variant is straight forward and we proceed to show how the effective reproduction number for a new variant can be estimated without contemporary sequencing results. This is useful for assessing the state of the pandemic in real time as we illustrate empirically with the inferred effective reproduction number for the Alpha variant.
翻译:我们提出一个简单的动态模型,用于估计两种病毒变异的相对传染性。最大可能性估计和推论容易使抽样期间的病例总数变化不定,并且可以作为一种后勤倒退来表示。我们将模型应用于丹麦SARS-COV-2变异数据。我们估计阿尔法和德尔塔的复制数比祖传变异的复制数大1.51倍[CI95%:1.50,1.53]和3.28 [CI95%:3.01,3.58]。在大多数接种疫苗的人口中,我们估计Omicron的传染性比德尔塔高出3.15倍[CI95%:2.83,3.50]。预测新出现的病毒变异的比例是直接向前的,我们继续显示如何在没有当代测序结果的情况下能够估计新变异的有效繁殖数。这对实时评估大流行病状况很有用,因为我们用阿尔法变异的推断有效繁殖数来进行实验。