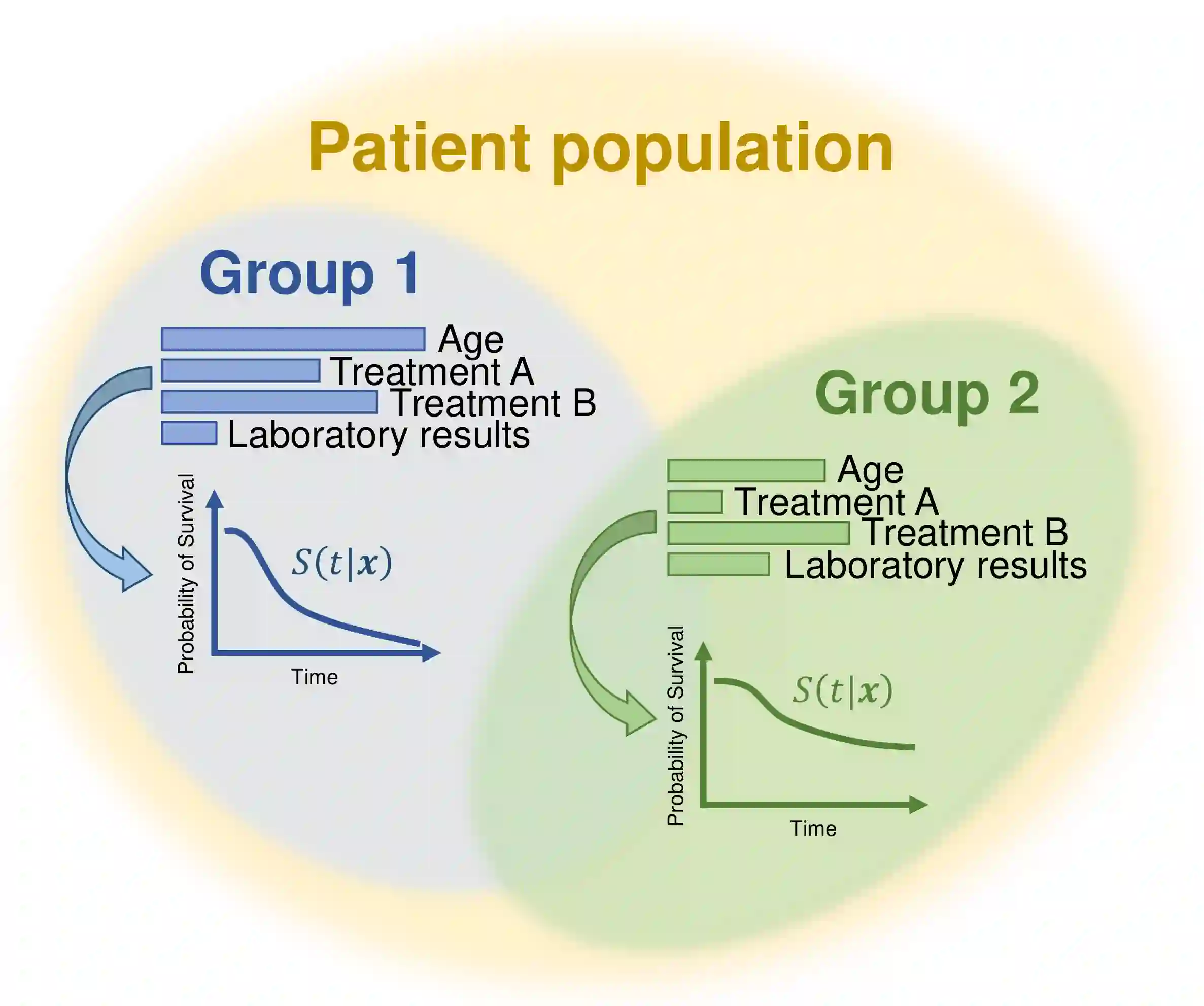
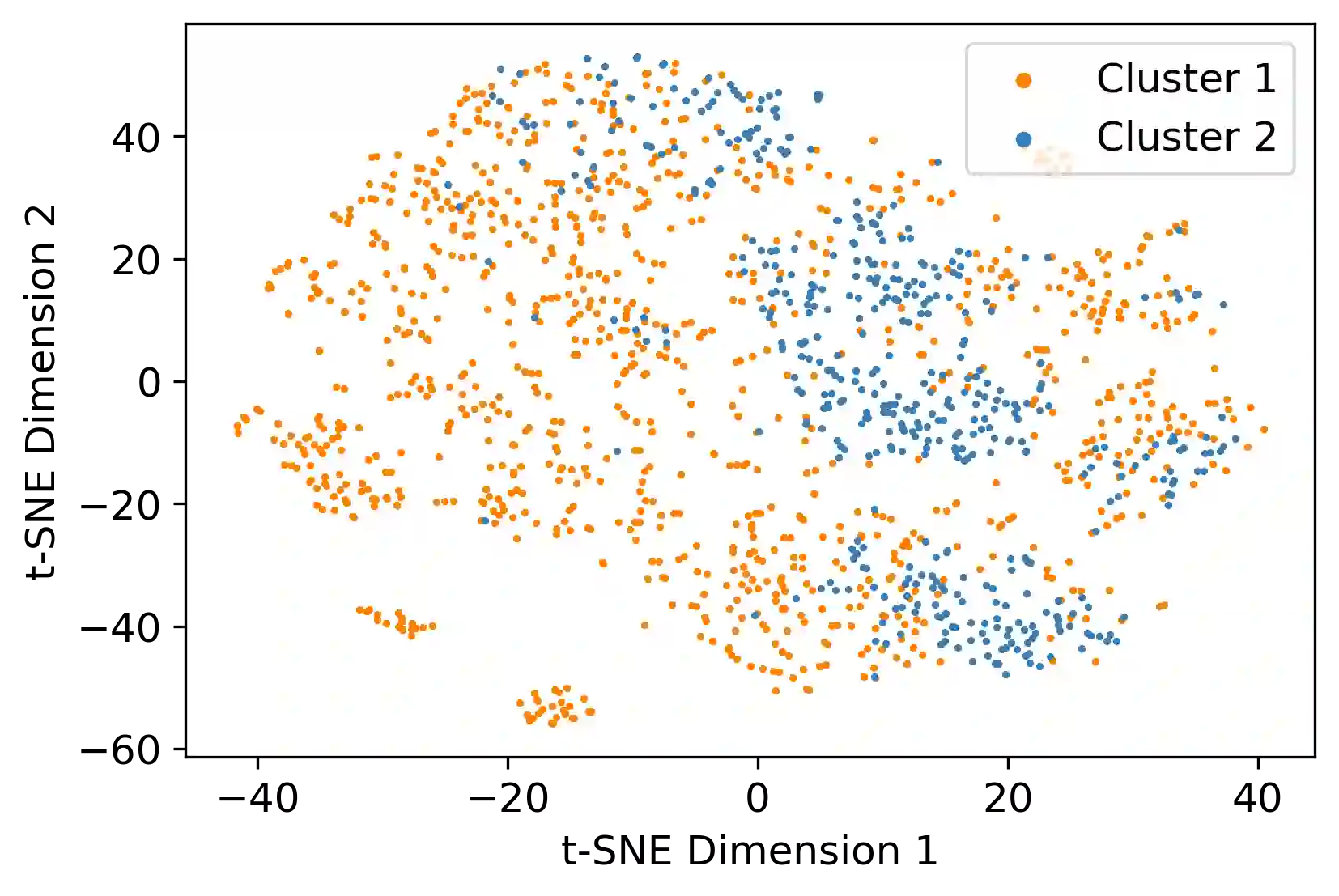
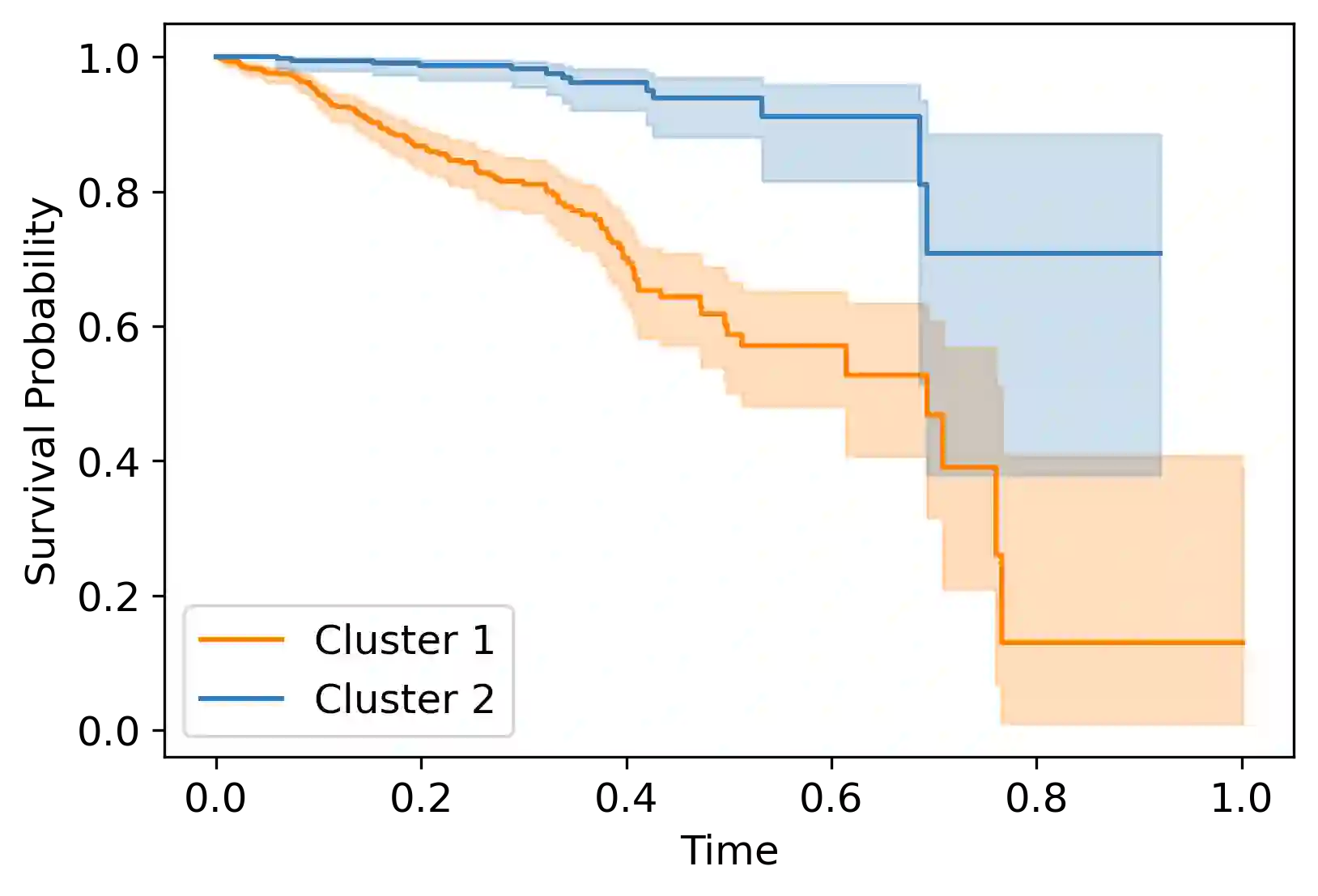
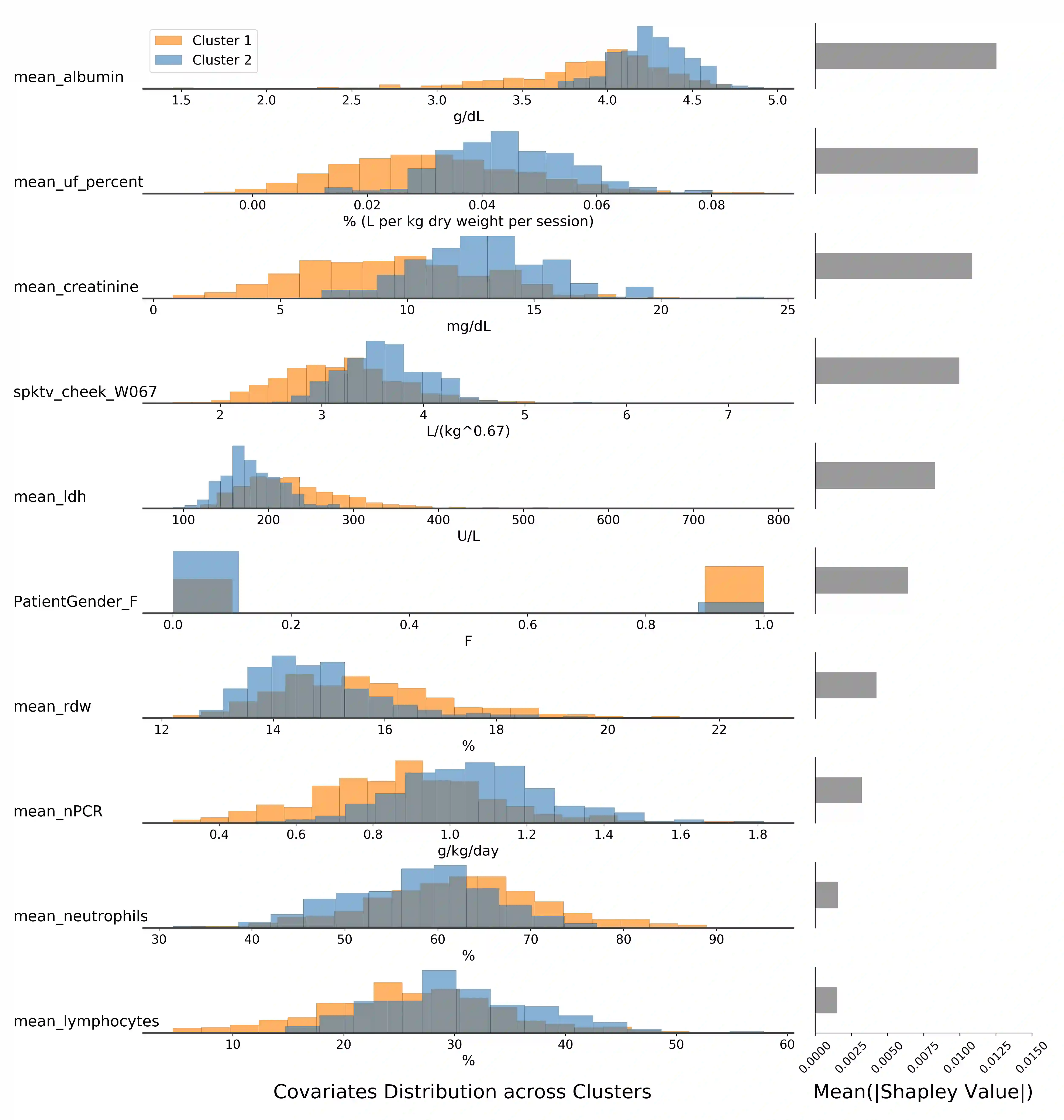
Survival analysis has gained significant attention in the medical domain and has many far-reaching applications. Although a variety of machine learning methods have been introduced for tackling time-to-event prediction in unstructured data with complex dependencies, clustering of survival data remains an under-explored problem. The latter is particularly helpful in discovering patient subpopulations whose survival is regulated by different generative mechanisms, a critical problem in precision medicine. To this end, we introduce a novel probabilistic approach to cluster survival data in a variational deep clustering setting. Our proposed method employs a deep generative model to uncover the underlying distribution of both the explanatory variables and the potentially censored survival times. We compare our model to the related work on survival clustering in comprehensive experiments on a range of synthetic, semi-synthetic, and real-world datasets. Our proposed method performs better at identifying clusters and is competitive at predicting survival times in terms of the concordance index and relative absolute error. To further demonstrate the usefulness of our approach, we show that our method identifies meaningful clusters from an observational cohort of hemodialysis patients that are consistent with previous clinical findings.
翻译:生存分析在医学领域受到极大关注,并具有许多深远的应用。虽然在复杂依赖性的非结构化数据中采用了各种机器学习方法来处理时间到活动预测,但生存数据组仍然是探索不足的问题,后者特别有助于发现由不同基因化机制管理生存的病人亚群,这是精确医学中的一个关键问题。为此,我们引入了一种新型的概率方法,在一个变异的深度集群环境中将生存数据组集成为生存数据组。我们提议的方法采用一种深层次的遗传模型,以发现解释变量和可能受审查的生存时间的根本分布。我们将我们的方法与综合实验中与一系列合成、半合成和现实世界数据集有关的生存群集工作进行比较。我们提议的方法在确定群集方面表现得更好,而且从一致性指数和相对绝对错误的角度预测生存时间具有竞争力。为了进一步证明我们的方法的有用性,我们的方法从与以往临床调查结果一致的热感解病人观察群中找出有意义的集群。