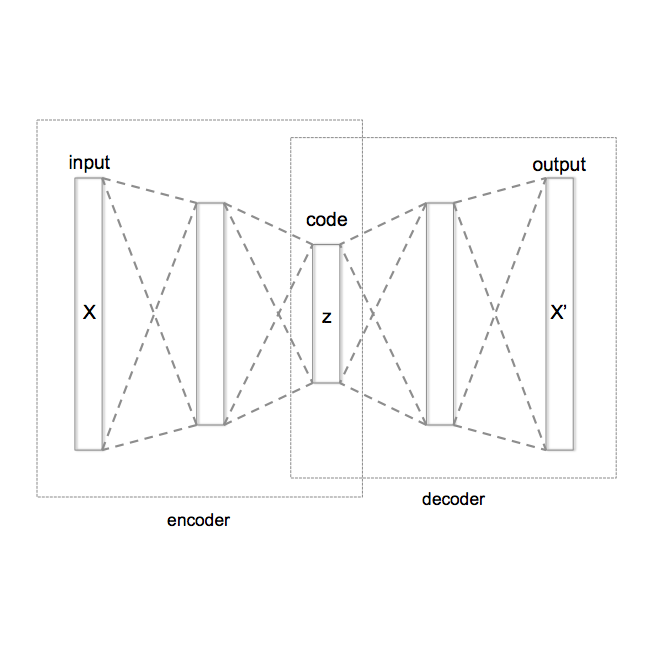
The lack of explainability is one of the most prominent disadvantages of deep learning applications in omics. This "black box" problem can undermine the credibility and limit the practical implementation of biomedical deep learning models. Here we present XOmiVAE, a variational autoencoder (VAE) based interpretable deep learning model for cancer classification using high-dimensional omics data. XOmiVAE is capable of revealing the contribution of each gene and latent dimension for each classification prediction, and the correlation between each gene and each latent dimension. It is also demonstrated that XOmiVAE can explain not only the supervised classification but the unsupervised clustering results from the deep learning network. To the best of our knowledge, XOmiVAE is one of the first activation level-based interpretable deep learning models explaining novel clusters generated by VAE. The explainable results generated by XOmiVAE were validated by both the performance of downstream tasks and the biomedical knowledge. In our experiments, XOmiVAE explanations of deep learning based cancer classification and clustering aligned with current domain knowledge including biological annotation and academic literature, which shows great potential for novel biomedical knowledge discovery from deep learning models.
翻译:缺乏解释性是渗透学中深层学习应用的最显著缺点之一。 这个“黑盒”问题不仅会破坏生物医学深层学习模型的可信度,还会限制实际应用。 我们在这里展示了基于变异自动读数器(VAE)的基于可解释的癌症分类的深层学习模型(VAE),用于使用高维显微镜数据进行癌症分类。 XOMIVAE能够揭示每种基因和潜在层面对分类预测的贡献,以及每个基因和每个潜在层面的关联性。它还表明 XOMIVAE不仅可以解释监督分类,而且可以解释深层次学习网络的未受监督的组合结果。根据我们的最佳知识, XOMIVAE是第一个基于不同层次的可解释性深层学习模型之一。 XOMIVAE产生的可解释性结果得到了下游任务表现和生物医学知识的验证。 在我们的实验中, XOMIVAE解释基于深度学习的癌症分类和组合与当前域知识(包括生物说明和学术文献)相一致的深层次知识的深度学习发现的巨大潜力。