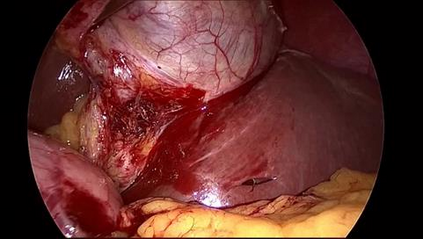
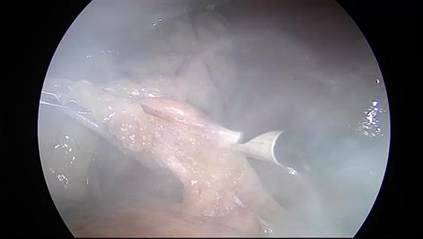
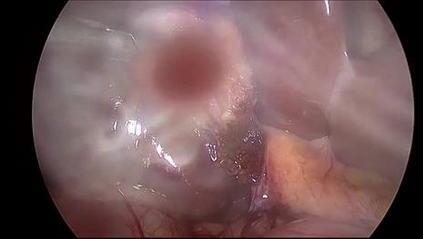
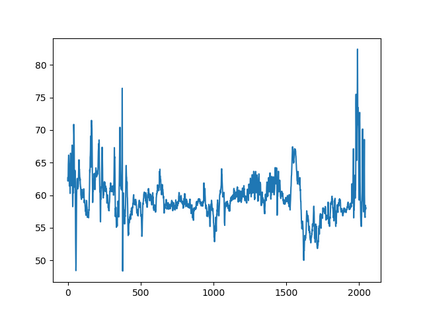
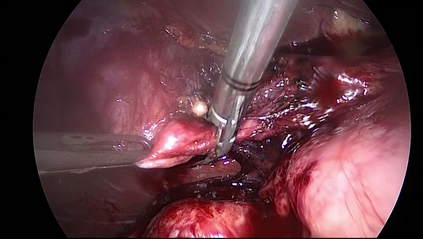
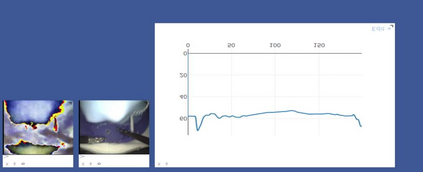
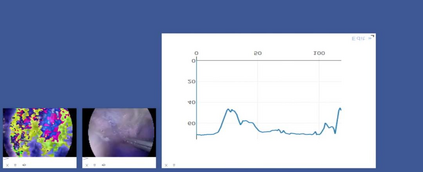
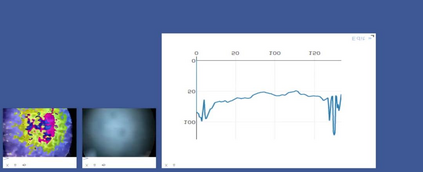
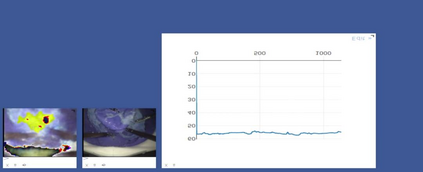
Anomaly detection in Minimally-Invasive Surgery (MIS) traditionally requires a human expert monitoring the procedure from a console. Data scarcity, on the other hand, hinders what would be a desirable migration towards autonomous robotic-assisted surgical systems. Automated anomaly detection systems in this area typically rely on classical supervised learning. Anomalous events in a surgical setting, however, are rare, making it difficult to capture data to train a detection model in a supervised fashion. In this work we thus propose an unsupervised approach to anomaly detection for robotic-assisted surgery based on deep residual autoencoders. The idea is to make the autoencoder learn the 'normal' distribution of the data and detect abnormal events deviating from this distribution by measuring the reconstruction error. The model is trained and validated upon both the publicly available Cholec80 dataset, provided with extra annotation, and on a set of videos captured on procedures using artificial anatomies ('phantoms') produced as part of the Smart Autonomous Robotic Assistant Surgeon (SARAS) project. The system achieves recall and precision equal to 78.4%, 91.5%, respectively, on Cholec80 and of 95.6%, 88.1% on the SARAS phantom dataset. The end-to-end system was developed and deployed as part of the SARAS demonstration platform for real-time anomaly detection with a processing time of about 25 ms per frame.
翻译:在小型侵入性外科手术(MIS)中异常检测传统上需要由人类专家从控制台对程序进行监测。另一方面,数据稀缺妨碍了向自主机器人辅助外科手术系统进行理想的迁移。这一领域的自动异常检测系统通常依赖古典监督的学习。但是,外科手术环境中异常事件很少,因此难以以监督的方式获取数据以培训检测模型。在这项工作中,我们提议对机器人辅助手术的异常检测采取不受监督的方法,该手术以深效自动识数仪为基础。其想法是让自动编码器学习数据“正常”的分布,并通过测量重建错误来探测从这一分布中偏离的异常事件。该模型在公开提供的Choolec80数据集(提供额外说明)和一套关于人工解剖仪(“假图”)程序采集的录像上得到培训和验证。在智能自主机器人助理外科医生(SAAS)平台(SARAS)项目中,SAS系统在SAR-88-1%的检测框架中,在SAR-85-1%的终端中,C-ral-ral-ral-ral-ral-ral-rental AS-ral-ral-ral-e-ral-ral-ral-ral-e-ral-ration-e-ral-e-a-e-e-e-s-sel-sex-sex-s-s-sex-s-s-s-s-s-s-s-s-s-s-s-s-s-s-s-s-s-xxxxxx-x-x-xxxxxx-x-x-xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx-x-x-x-x-x-x-x-xxxxx-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-x-xx-x-x-x-x-x-x-x-