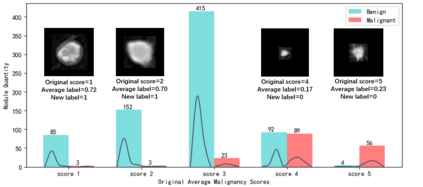
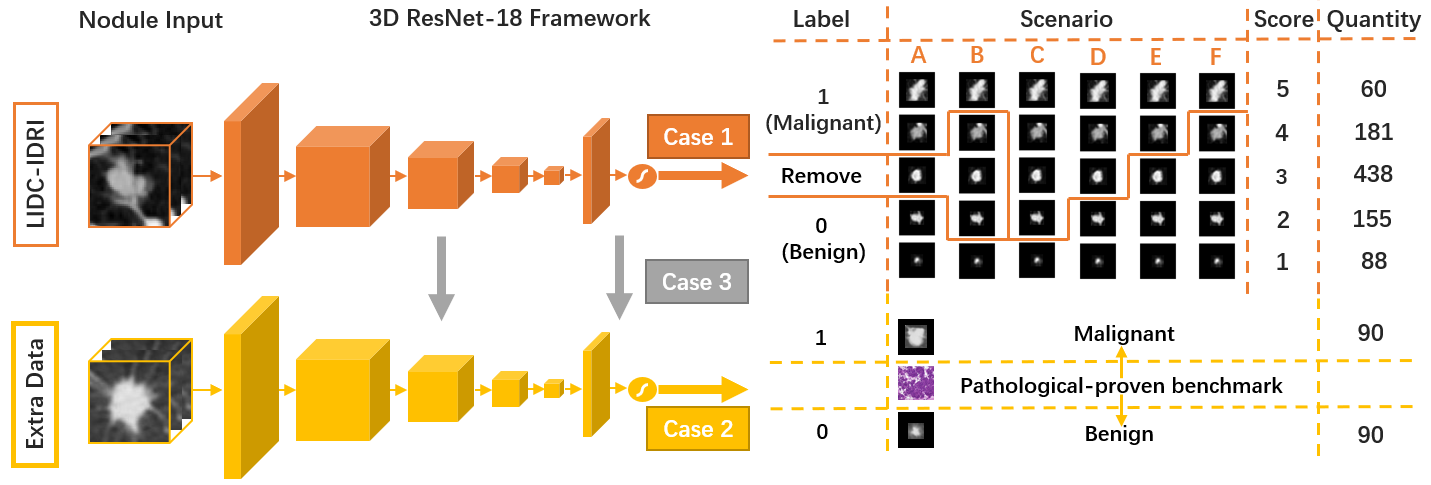
The LIDC-IDRI database is the most popular benchmark for lung cancer prediction. However, with subjective assessment from radiologists, nodules in LIDC may have entirely different malignancy annotations from the pathological ground truth, introducing label assignment errors and subsequent supervision bias during training. The LIDC database thus requires more objective labels for learning-based cancer prediction. Based on an extra small dataset containing 180 nodules diagnosed by pathological examination, we propose to re-label LIDC data to mitigate the effect of original annotation bias verified on this robust benchmark. We demonstrate in this paper that providing new labels by similar nodule retrieval based on metric learning would be an effective re-labeling strategy. Training on these re-labeled LIDC nodules leads to improved model performance, which is enhanced when new labels of uncertain nodules are added. We further infer that re-labeling LIDC is current an expedient way for robust lung cancer prediction while building a large pathological-proven nodule database provides the long-term solution.
翻译:LIDDC-IDRI数据库是肺癌预测最受欢迎的基准。然而,根据放射学家的主观评估,LIDC结核的恶性说明可能完全不同于病理地面真理,在培训期间引入标签分配错误和随后的监督偏差。因此,LIDC数据库需要更客观的癌症学习预测标签。根据由病理检查诊断的包含180个结核的额外小数据集,我们提议重新标签LIDC数据,以减轻根据这一稳健基准核实的原始说明偏差的影响。我们在本文件中表明,通过类似的结核检索提供新的标签,将是一种有效的再标签战略。关于这些重新标签的LIDC结核的培训可以改进模型性能,在添加不确定结核的新标签时,这种能力会得到加强。我们进一步推想,再标签LIDC目前是稳健的肺癌预测的方便方法,同时建立一个大型病理学预测结核数据库,提供长期的解决办法。