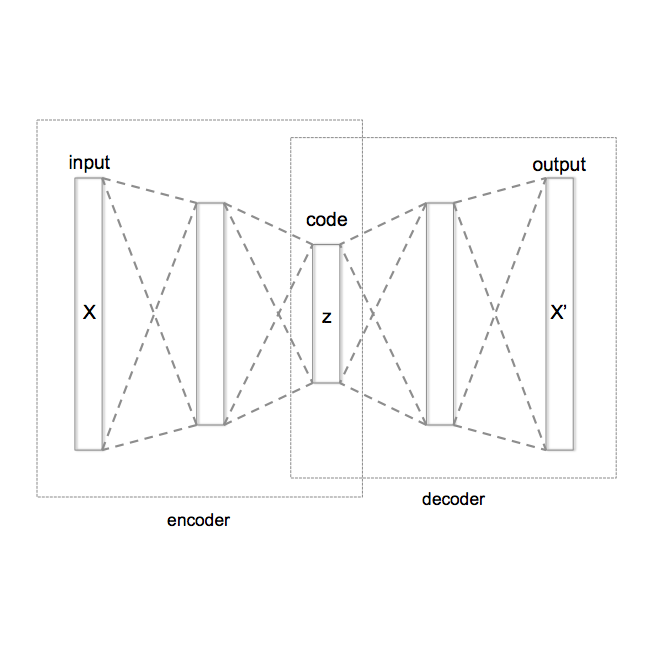
We develop the sparse VAE for unsupervised representation learning on high-dimensional data. The sparse VAE learns a set of latent factors (representations) which summarize the associations in the observed data features. The underlying model is sparse in that each observed feature (i.e. each dimension of the data) depends on a small subset of the latent factors. As examples, in ratings data each movie is only described by a few genres; in text data each word is only applicable to a few topics; in genomics, each gene is active in only a few biological processes. We prove such sparse deep generative models are identifiable: with infinite data, the true model parameters can be learned. (In contrast, most deep generative models are not identifiable.) We empirically study the sparse VAE with both simulated and real data. We find that it recovers meaningful latent factors and has smaller heldout reconstruction error than related methods.
翻译:我们开发了稀疏的 VAE, 用于在高维数据上进行不受监督的演示学习。 稀疏的 VAE 学习了一组潜在因素( 代表), 概括了观察到的数据特征中的关联性。 基本模型很稀少, 因为每个观察到的特征( 即数据的每个维度) 都取决于一小部分潜在因素。 例如, 在评级数据中, 每一部电影只用几小类来描述; 在文本数据中, 每个单词都只适用于几个主题; 在基因组学中, 每个基因只活跃在几个生物过程中。 我们证明这种稀疏的深深层基因模型是可识别的: 有了无限的数据, 真正的模型参数是可以学的 。 ( 相比之下, 大部分深厚的基因模型是无法识别的 ) 我们用模拟和真实的数据对稀疏的 VAE 进行实验性研究。 我们发现, 它恢复了有意义的潜在因素, 并且比相关方法要少一些隐藏的重建错误 。