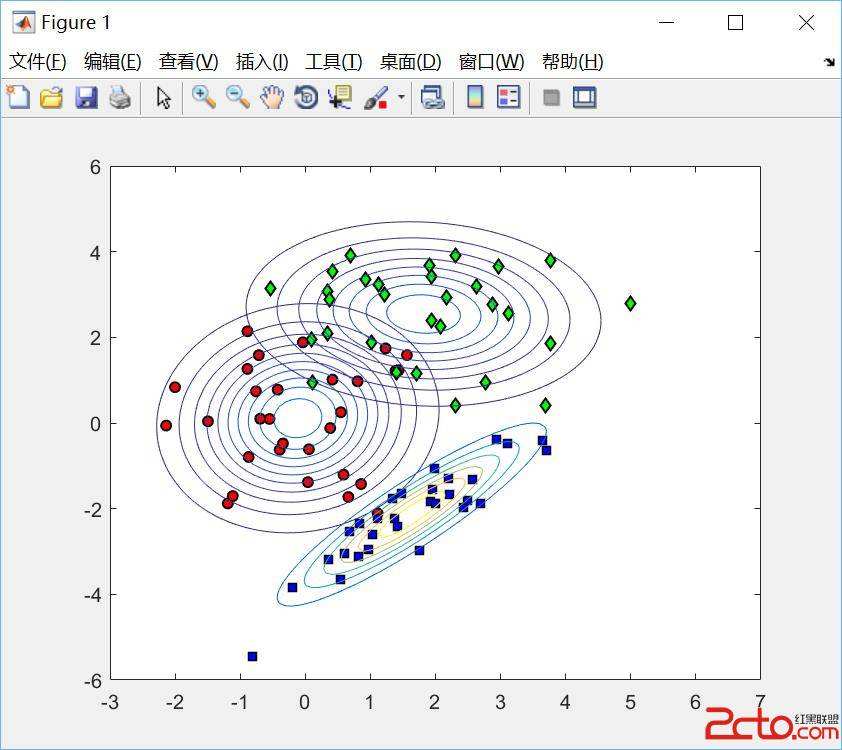
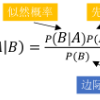
Whether based on models, training data or a combination, classifiers place (possibly complex) input data into one of a relatively small number of output categories. In this paper, we study the structure of the boundary--those points for which a neighbor is classified differently--in the context of an input space that is a graph, so that there is a concept of neighboring inputs, The scientific setting is a model-based naive Bayes classifier for DNA reads produced by Next Generation Sequencers. We show that the boundary is both large and complicated in structure. We create a new measure of uncertainty, called Neighbor Similarity, that compares the result for a point to the distribution of results for its neighbors. This measure not only tracks two inherent uncertainty measures for the Bayes classifier, but also can be implemented, at a computational cost, for classifiers without inherent measures of uncertainty.
翻译:暂无翻译