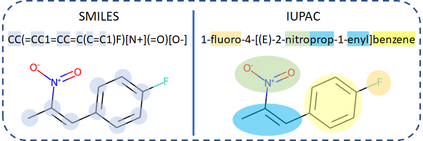
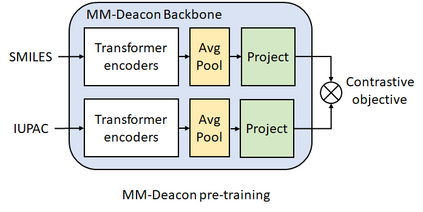
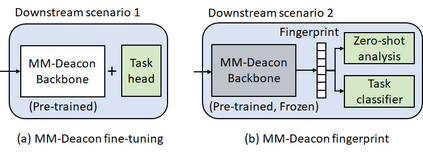
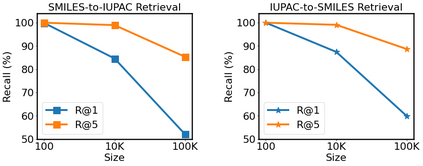
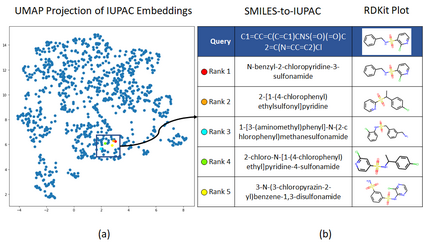
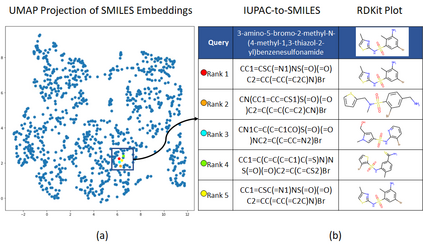
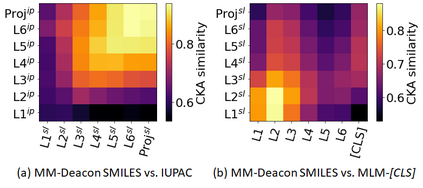
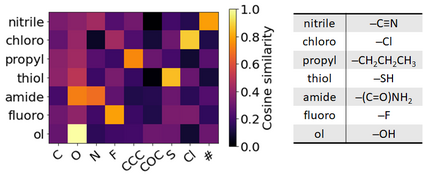
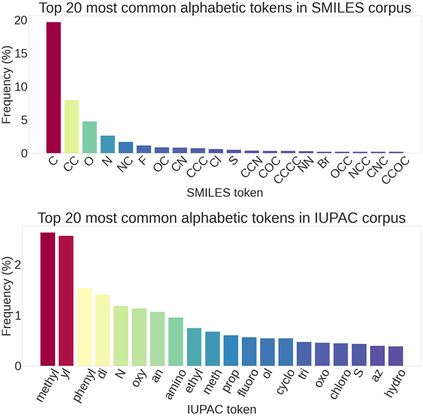
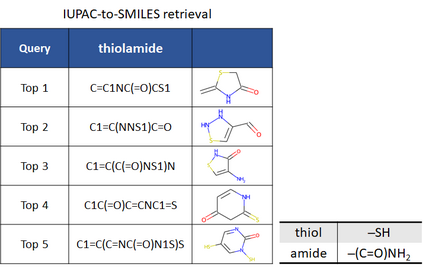
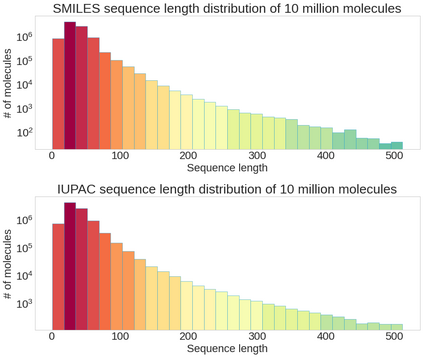
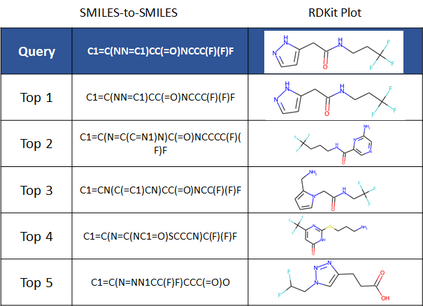
Molecular representation learning plays an essential role in cheminformatics. Recently, language model-based approaches have gained popularity as an alternative to traditional expert-designed features to encode molecules. However, these approaches only utilize a single molecular language for representation learning. Motivated by the fact that a given molecule can be described using different languages such as Simplified Molecular Line Entry System (SMILES), The International Union of Pure and Applied Chemistry (IUPAC), and The IUPAC International Chemical Identifier (InChI), we propose a multilingual molecular embedding generation approach called MM-Deacon (multilingual molecular domain embedding analysis via contrastive learning). MM-Deacon is pre-trained using SMILES and IUPAC as two different languages on large-scale molecules. We evaluated the robustness of our method on seven molecular property prediction tasks from MoleculeNet benchmark, zero-shot cross-lingual retrieval, and a drug-drug interaction prediction task.
翻译:分子代表性学习在化学信息学中发挥着必不可少的作用。最近,语言模型方法作为传统专家设计的分子编码特征的替代方法,越来越受欢迎,不过,这些方法只使用单一分子语言进行代言学习,其动机是使用简化分子线输入系统(SMILES)、国际理论和应用化学联合会(IUPAC)和IUPAC国际化学识别仪(InCHI)等不同语言来描述特定分子,我们建议采用多语言分子嵌入代用方法,称为MM-Deacon(通过对比性学习进行多语言分子域嵌入分析)。MM-Deacon是预先培训的,使用SMILES和国际理论分子存取中心(IUPAC)作为两种关于大型分子的不同语言。我们评估了我们从MoleculeNet基准、零速跨语言检索和药物-药物互动预测任务中进行七种分子属性预测的方法是否健全。