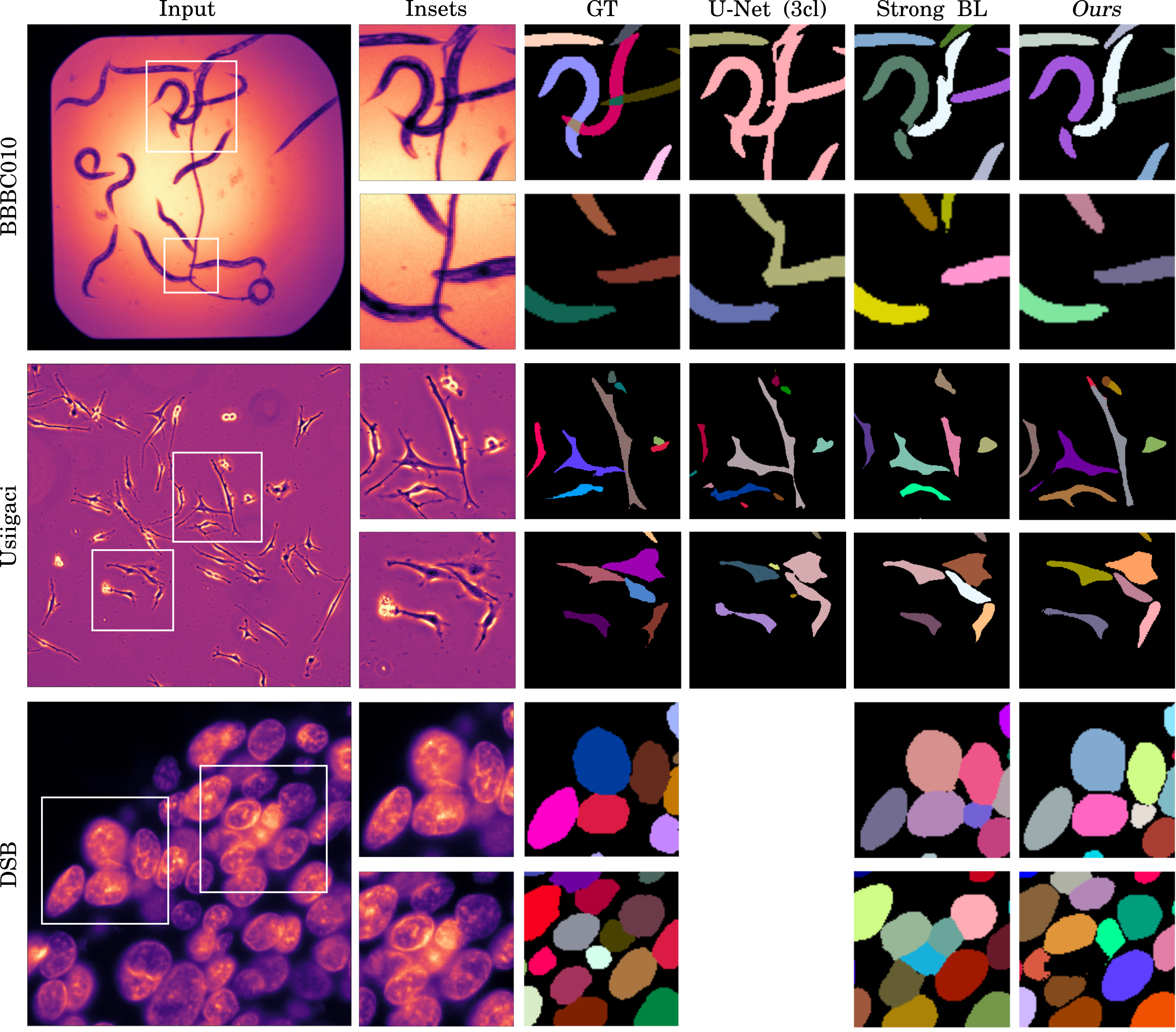
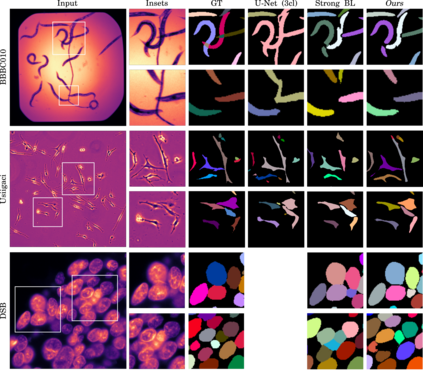
Automatic detection and segmentation of objects in microscopy images is important for many biological applications. In the domain of natural images, and in particular in the context of city street scenes, embedding-based instance segmentation leads to high-quality results. Inspired by this line of work, we introduce EmbedSeg, an end-to-end trainable deep learning method based on the work by Neven et al. While their approach embeds each pixel to the centroid of any given instance, in EmbedSeg, motivated by the complex shapes of biological objects, we propose to use the medoid instead. Additionally, we make use of a test-time augmentation scheme, and show that both suggested modifications improve the instance segmentation performance on biological microscopy datasets notably. We demonstrate that embedding-based instance segmentation achieves competitive results in comparison to state-of-the-art methods on diverse and biologically relevant microscopy datasets. Finally, we show that the overall pipeline has a small enough memory footprint to be used on virtually all CUDA enabled laptop hardware. Our open-source implementation is available at github.com/juglab/EmbedSeg.
翻译:对于许多生物应用而言,显微镜图像中天体的自动检测和分解非常重要。在自然图像领域,特别是在城市街道景象方面,嵌入式样的分解导致高质量的结果。在这项工作的启发下,我们采用了基于Neven 等人所做工作的、端到端可训练的深层学习方法EmbedSeg。虽然它们的方法将每个像素嵌入任何特定例子的中位体,在EmbedSeg, 由生物物体的复杂形状所驱动,我们提议代之使用类。此外,我们采用测试-时间增强计划,并表明建议的两项修改都提高了生物显微镜数据集的分解性性能。我们证明,嵌入式样分解与关于多样化和生物相关显微镜数据集的先进方法相比,具有竞争性的结果。最后,我们表明整个管道的记忆足迹足迹可以用于几乎所有CUDA启用的膝上硬件。我们的开源实施工作在Github.Sjugrob.Srogla。