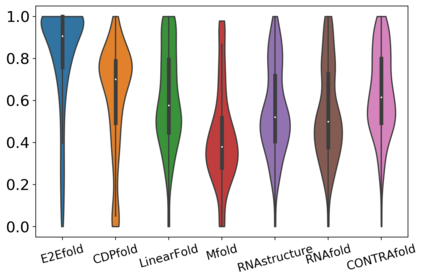
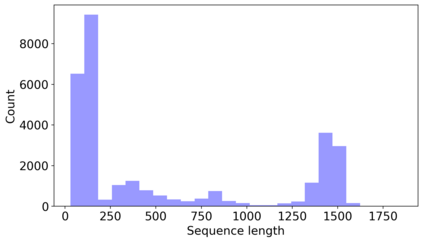
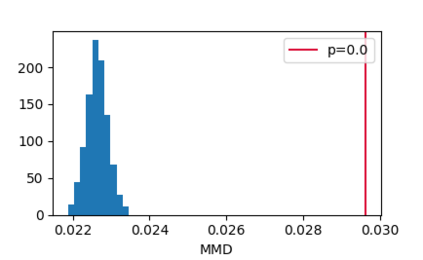
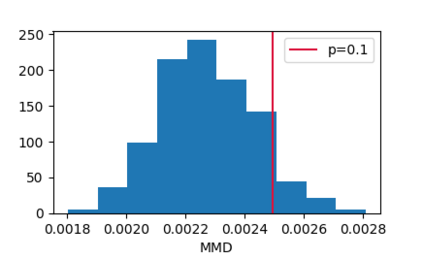
In this paper, we propose an end-to-end deep learning model, called E2Efold, for RNA secondary structure prediction which can effectively take into account the inherent constraints in the problem. The key idea of E2Efold is to directly predict the RNA base-pairing matrix, and use an unrolled algorithm for constrained programming as the template for deep architectures to enforce constraints. With comprehensive experiments on benchmark datasets, we demonstrate the superior performance of E2Efold: it predicts significantly better structures compared to previous SOTA (especially for pseudoknotted structures), while being as efficient as the fastest algorithms in terms of inference time.
翻译:在本文中,我们为RNA二级结构预测提出了一个端到端深学习模型,称为E2Efold,该模型可以有效地考虑到这一问题的内在限制。 E2Eform的关键想法是直接预测RNA基底涂料矩阵,并使用无动于衷的算法来限制编程,作为实施限制的深层结构模板。在对基准数据集进行全面实验后,我们展示了E2Eform的优异性能:它预测的结构比以前的SOTA(特别是假记结构)要好得多,同时在推算时间方面效率也和最快的算法一样高。