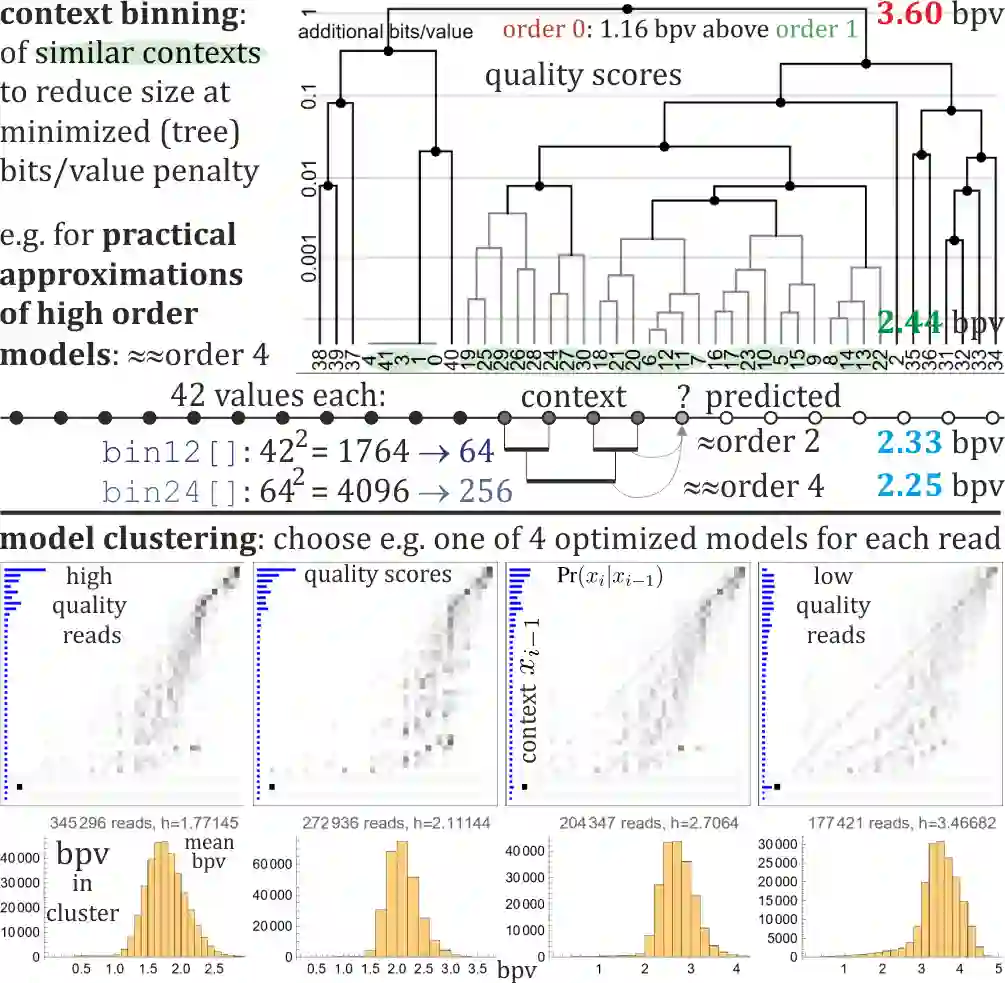
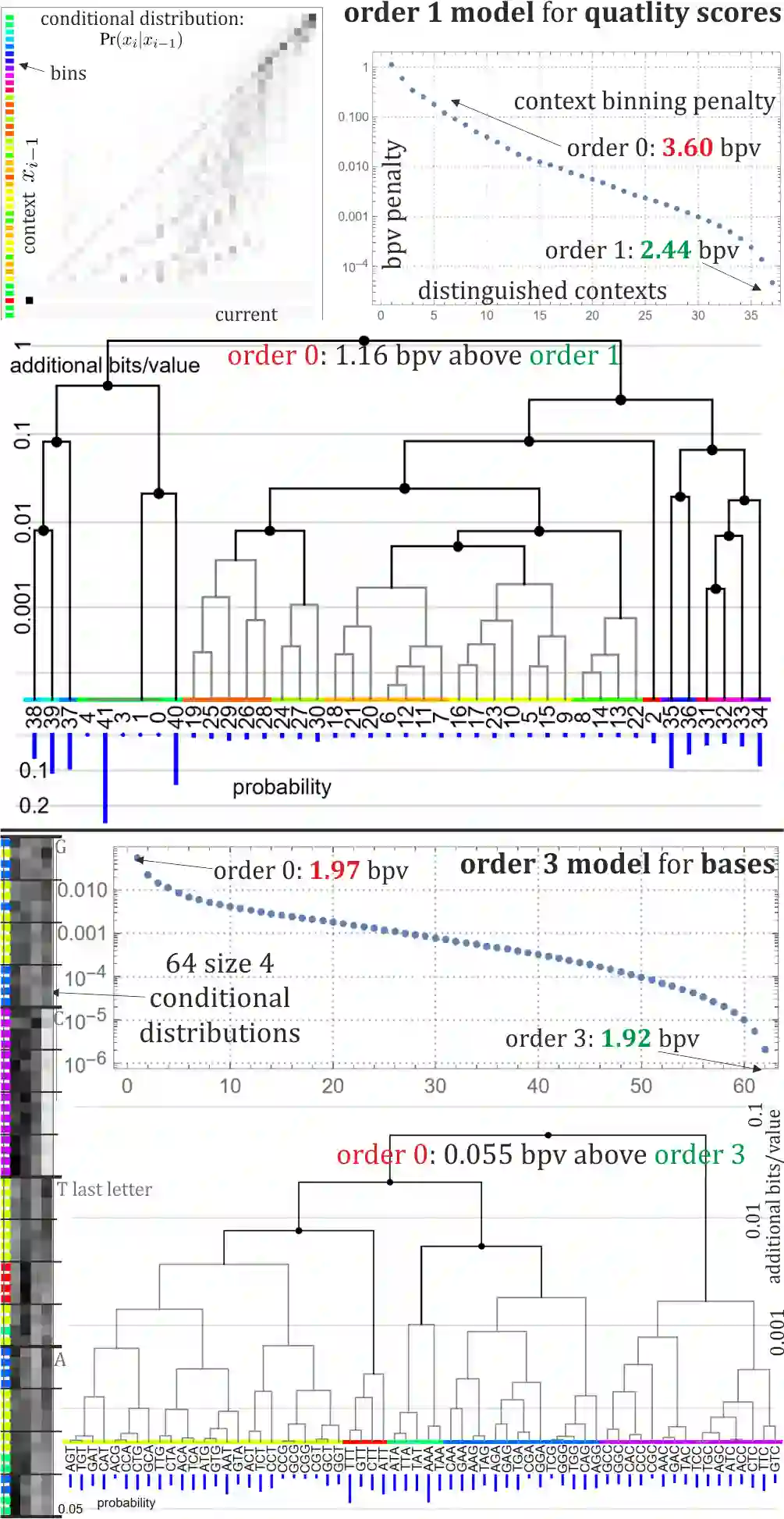
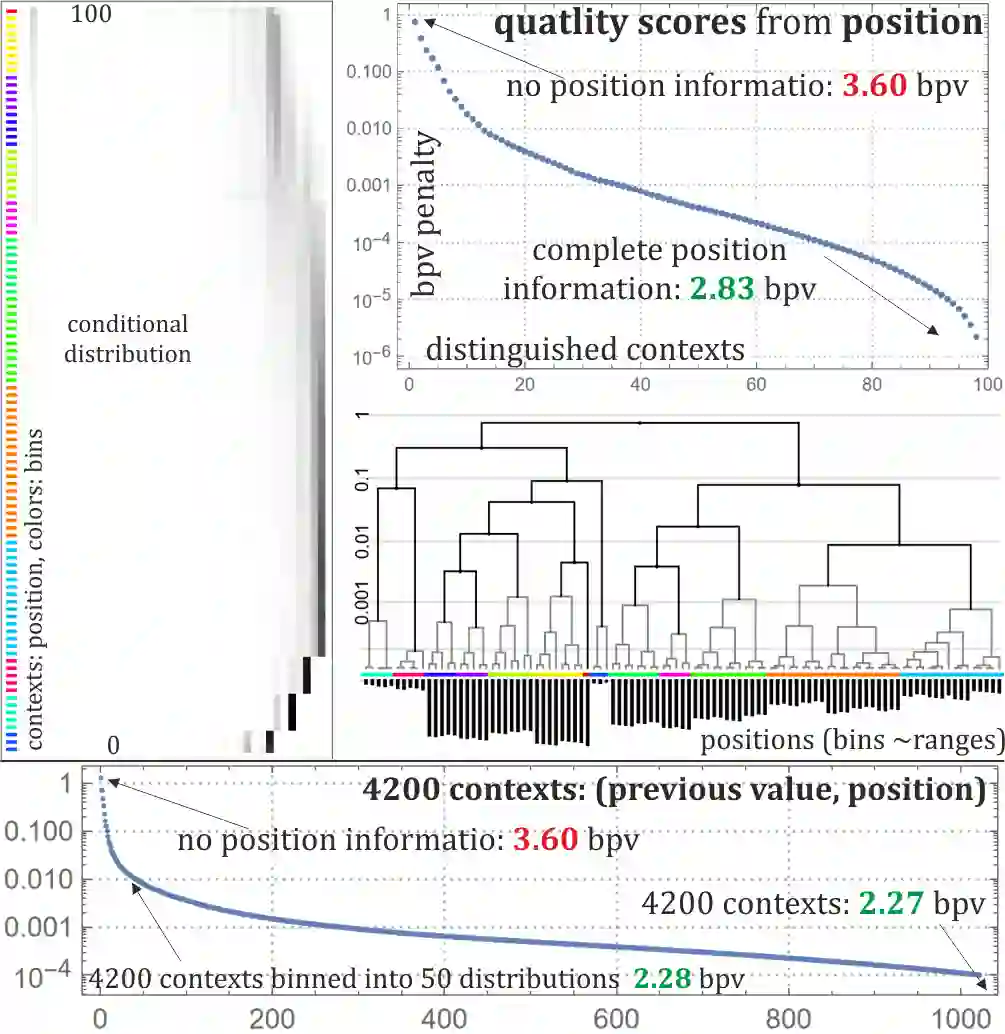
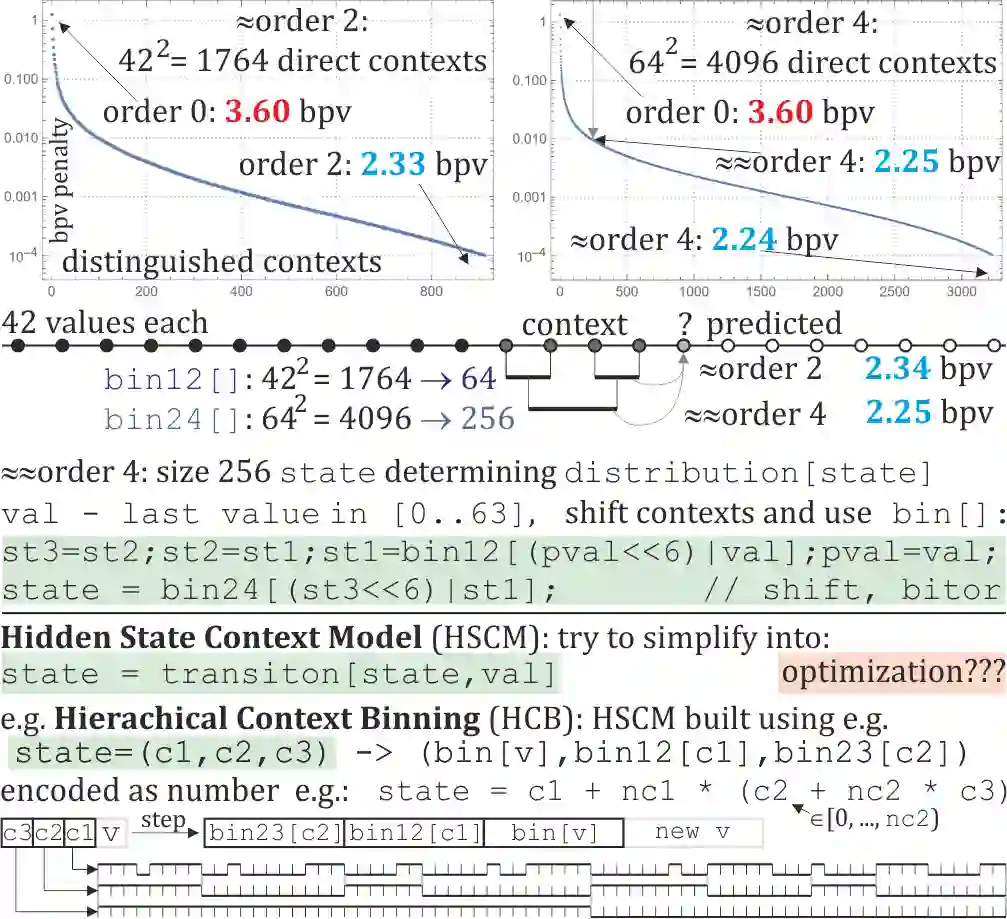
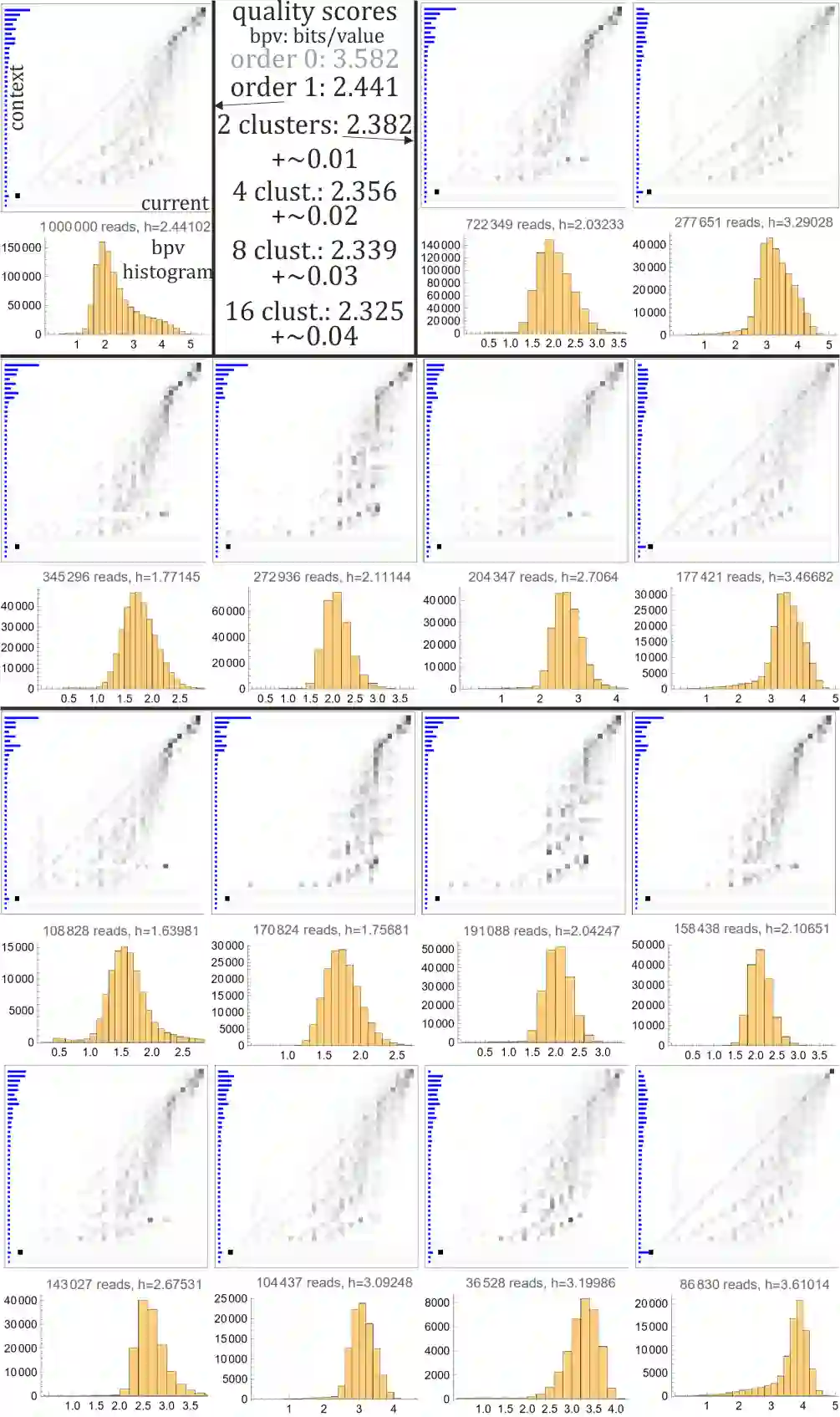
Rapid growth of genetic databases means huge savings from improvements in their data compression, what requires better inexpensive statistical models. This article proposes automatized optimizations e.g. of Markov-like models, especially context binning and model clustering. While it is popular to just remove low bits of the context, proposed context binning automatically optimizes such reduction as tabled: state=bin[context] determining probability distribution, this way extracting nearly all useful information also from very large contexts, into a relatively small number of states. The second proposed approach: model clustering uses k-means clustering in space of general statistical models, allowing to optimize a few models (as cluster centroids) to be chosen e.g. separately for each read. There are also briefly discussed some adaptivity techniques to include data non-stationarity.
翻译:基因数据库的快速增长意味着从数据压缩的改进中节省大量资金,这需要更廉价的统计模型。 本条提出自动优化, 如Markov相似的模型, 特别是环境拆迁和模型群集。 虽然只是删除低位环境位子很受欢迎, 拟议的环境拆迁会自动优化减排, 如: 国家=bin[Cext] 确定概率分布, 这样将几乎所有有用的信息也从非常大的背景中提取到相对较少的州。 第二种拟议方法: 模型群集在一般统计模型的空间中使用 k 手段群集, 以便优化选择的少数模型( 作为分类的中间体 ), 例如为每读取而分别选择 。 还简要讨论了一些适应性技术, 以包括数据非静态 。