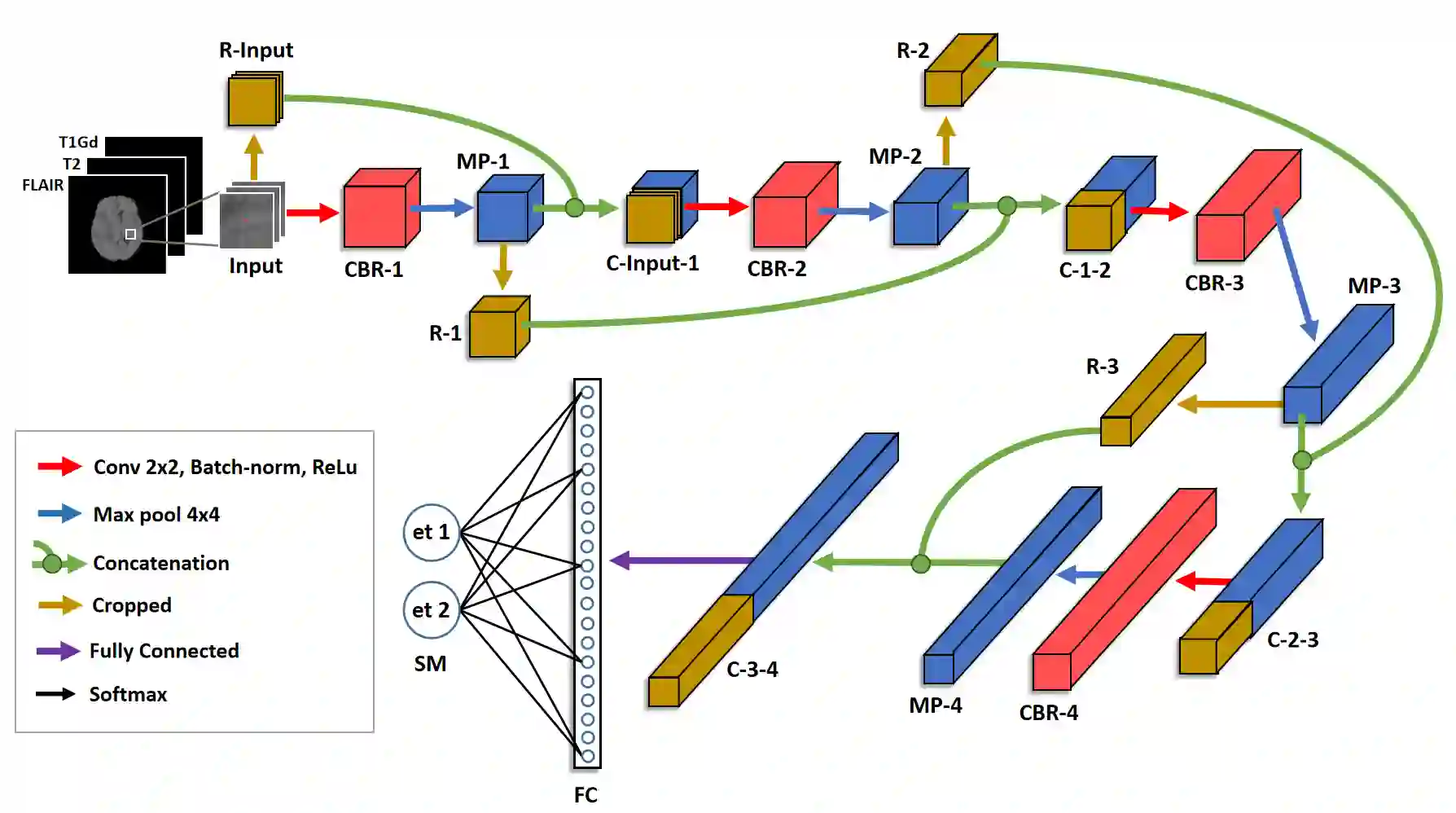
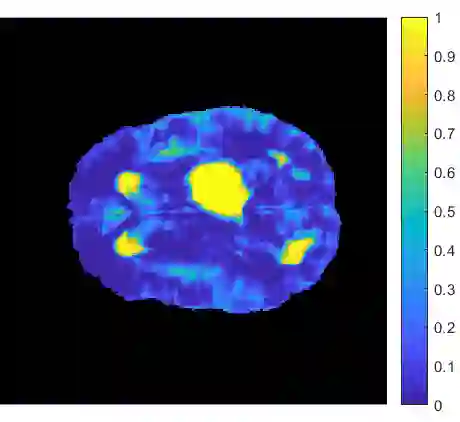
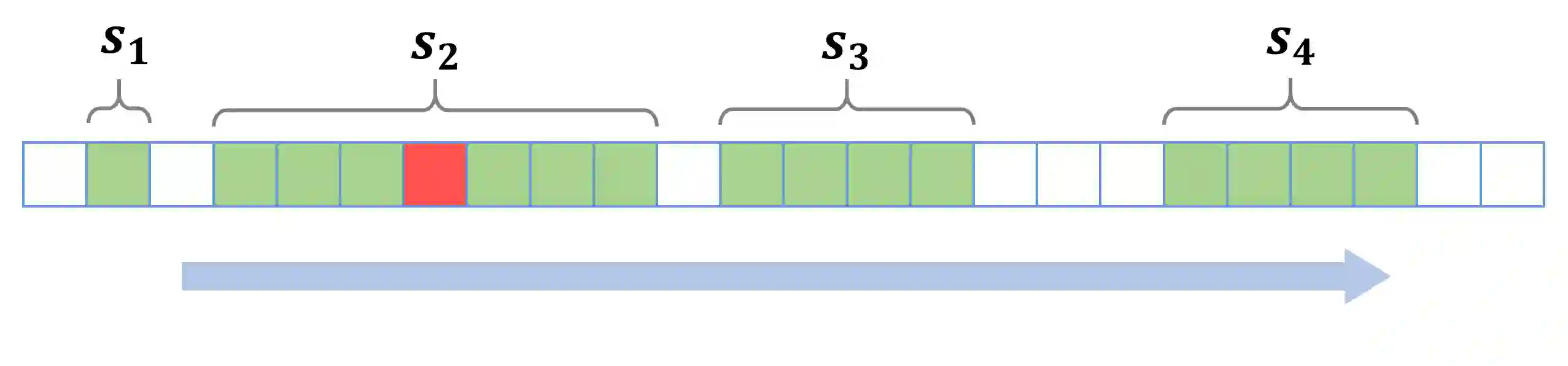
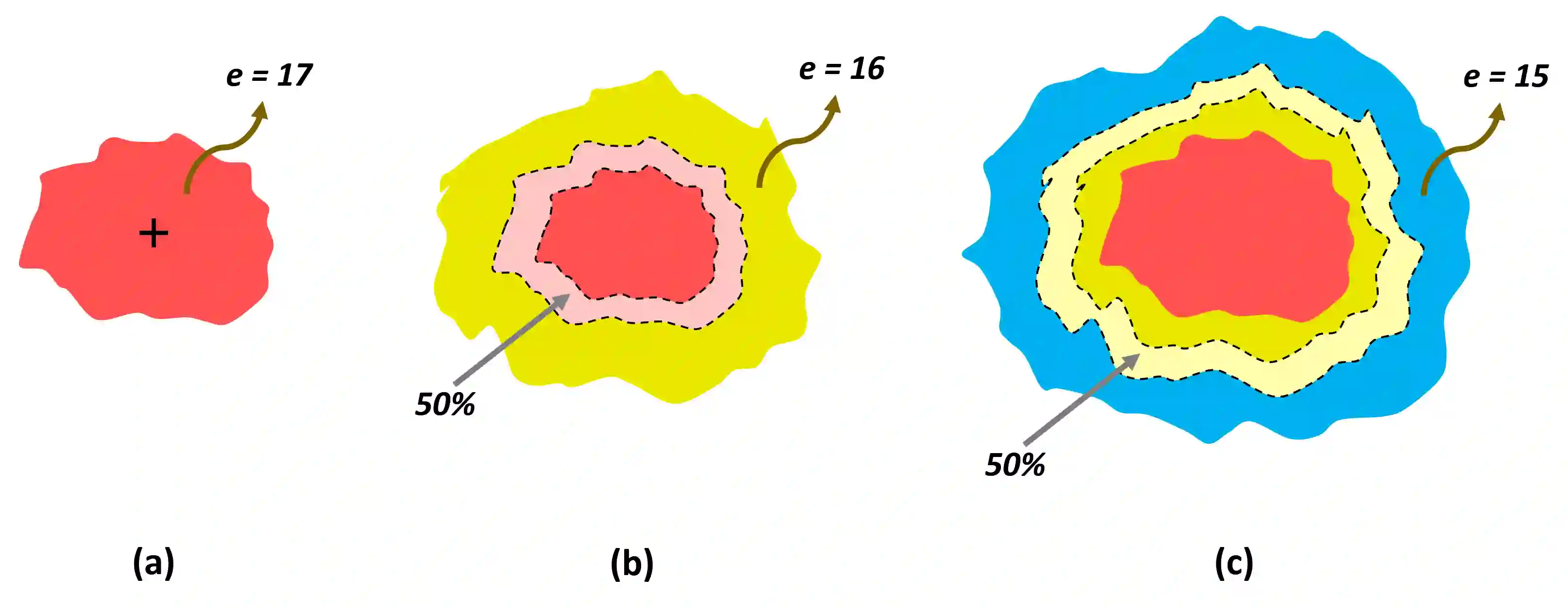
Gliomas are brain tumors composed of different highly heterogeneous histological subregions. Image analysis techniques to identify relevant tumor substructures have high potential for improving patient diagnosis, treatment and prognosis. However, due to the high heterogeneity of gliomas, the segmentation task is currently a major challenge in the field of medical image analysis. In the present work, the database of the Brain Tumor Segmentation (BraTS) Challenge 2018, composed of multimodal MRI scans of gliomas, was studied. A segmentation methodology based on the design and application of convolutional neural networks (CNNs) combined with original post-processing techniques with low computational demand was proposed. The post-processing techniques were the main responsible for the results obtained in the segmentations. The segmented regions were the whole tumor, the tumor core, and the enhancing tumor core, obtaining averaged Dice coefficients equal to 0.8934, 0.8376, and 0.8113, respectively. These results reached the state of the art in glioma segmentation determined by the winners of the challenge.
翻译:Gliomas是由不同高度多样化的神学次区域组成的脑肿瘤。通过图像分析技术,确定相关的肿瘤子结构,对于改善病人诊断、治疗和预测具有很大潜力。然而,由于微粒的异质性很高,分解任务目前是医学图像分析领域的一大挑战。在目前的工作中,对2018年脑肿瘤分解(BraTS)挑战数据库进行了研究,该数据库由对微粒的多式MRI扫描组成。根据超导神经网络的设计和应用,以及最初的后处理技术而计算需求低的分解方法,提出了后处理技术,主要负责分解结果。分解区域是整个肿瘤、肿瘤核心和增强的肿瘤核心,获得的平均骰子系数分别为0.8934、0.8376和0.8113。这些结果达到了挑战获胜者确定的液分解的艺术状态。