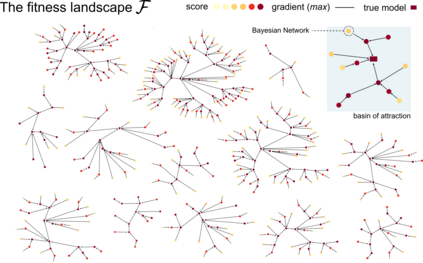
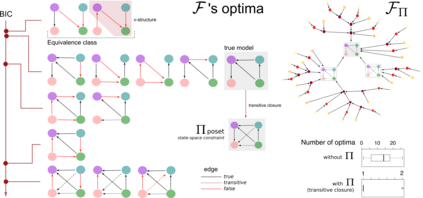
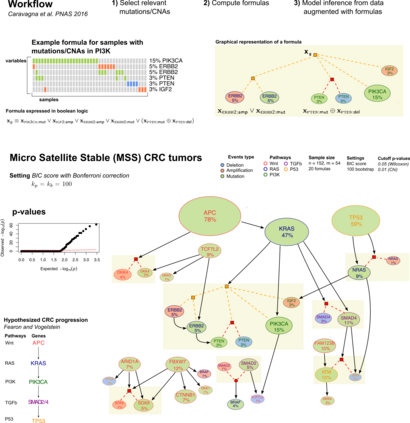
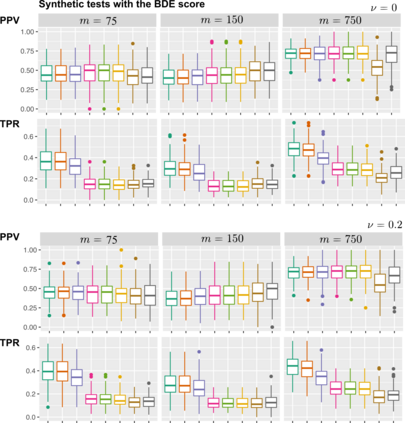
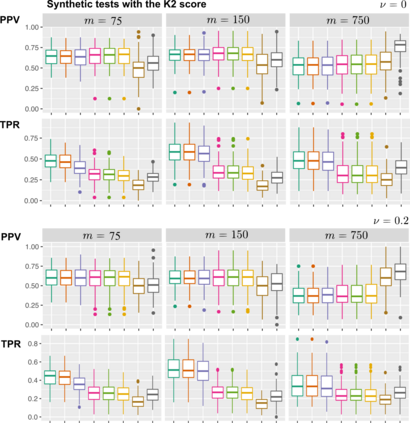
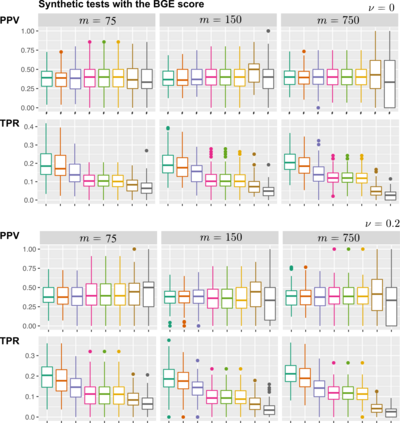
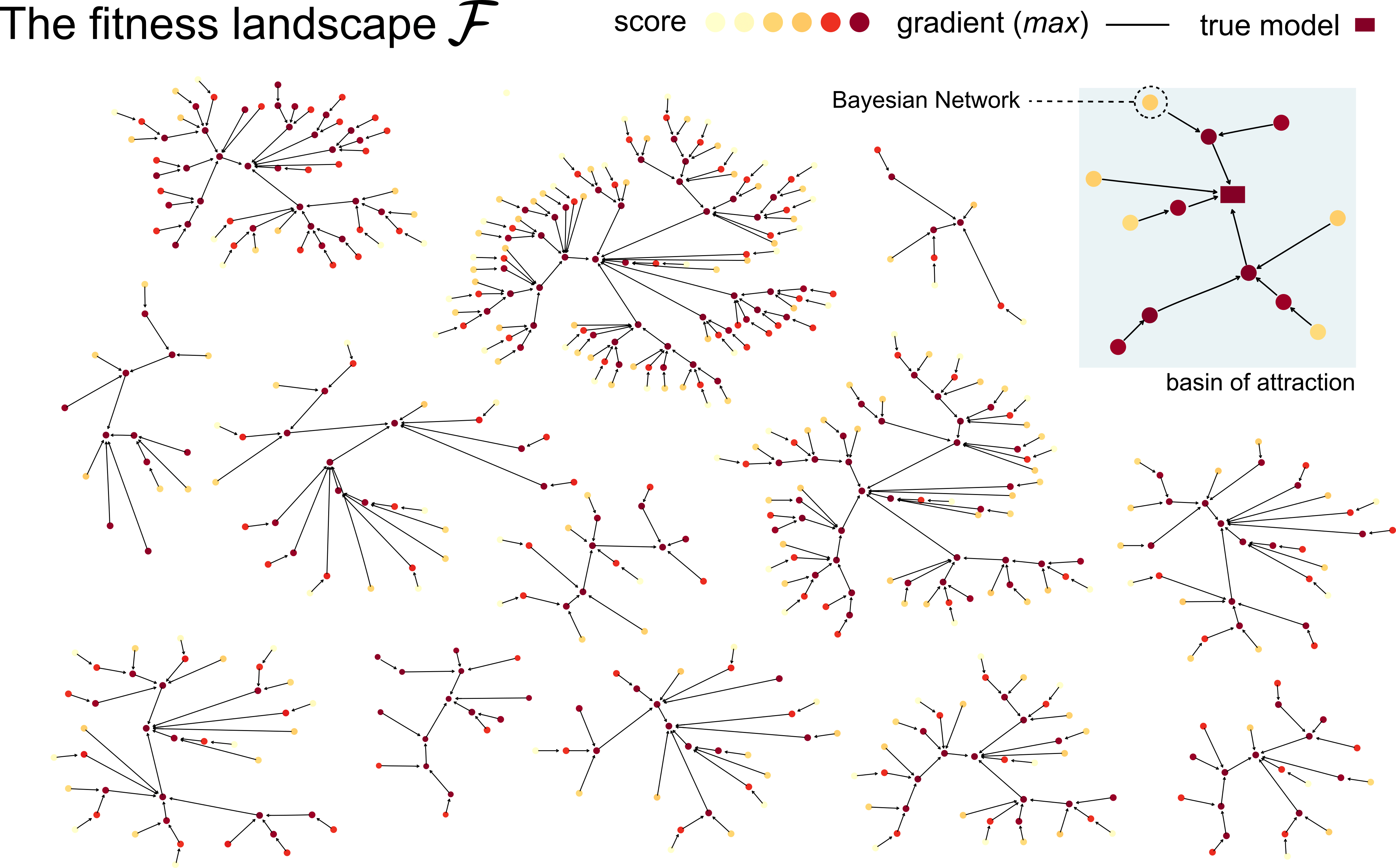
Learning the structure of dependencies among multiple random variables is a problem of considerable theoretical and practical interest. Within the context of Bayesian Networks, a practical and surprisingly successful solution to this learning problem is achieved by adopting score-functions optimisation schema, augmented with multiple restarts to avoid local optima. Yet, the conditions under which such strategies work well are poorly understood, and there are also some intrinsic limitations to learning the directionality of the interaction among the variables. Following an early intuition of Friedman and Koller, we propose to decouple the learning problem into two steps: first, we identify a partial ordering among input variables which constrains the structural learning problem, and then propose an effective bootstrap-based algorithm to simulate augmented data sets, and select the most important dependencies among the variables. By using several synthetic data sets, we show that our algorithm yields better recovery performance than the state of the art, increasing the chances of identifying a globally-optimal solution to the learning problem, and solving also well-known identifiability issues that affect the standard approach. We use our new algorithm to infer statistical dependencies between cancer driver somatic mutations detected by high-throughput genome sequencing data of multiple colorectal cancer patients. In this way, we also show how the proposed methods can shade new insights about cancer initiation, and progression. Code: https://github.com/caravagn/Bootstrap-based-Learning
翻译:在Bayesian 网络中,通过采用分数功能优化方案,并辅之以多次重新启动,以避免本地opima 选择。然而,这些战略在哪些条件下运作良好,而且在学习变量之间相互作用的方向性方面也存在一些内在限制。在Friedman和Koller的早期直觉之后,我们提议将学习问题分为两个步骤:首先,我们确定限制结构性学习问题的投入变量之间的部分排序,然后提出一种有效的基于靴杆的有效算法,以模拟扩大数据集,并在变量中选择最重要的依赖关系。我们通过使用几个合成数据集,表明我们的算法产生比艺术状态更好的恢复业绩,增加找出全球最佳学习问题解决方案的机会,并解决影响标准方法的众所周知的可识别性问题。我们用新的算法来推断多种统计依赖性、癌症患者/癌症患者的变异性。我们用高的算法来显示癌症变异性排序方法。