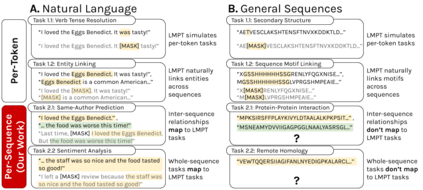
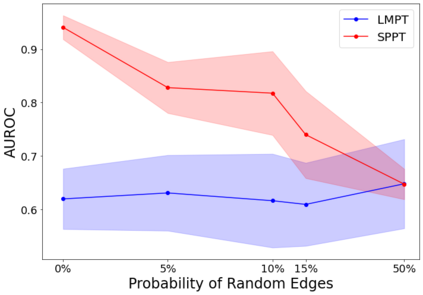
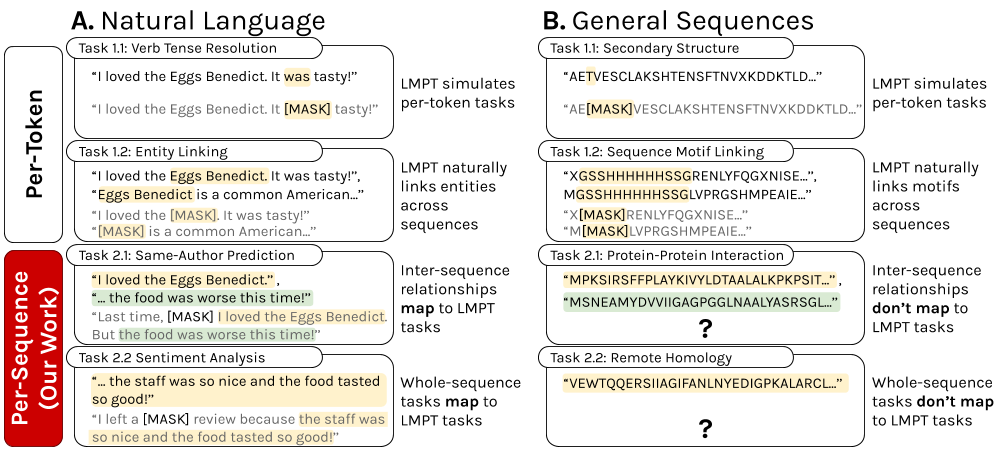
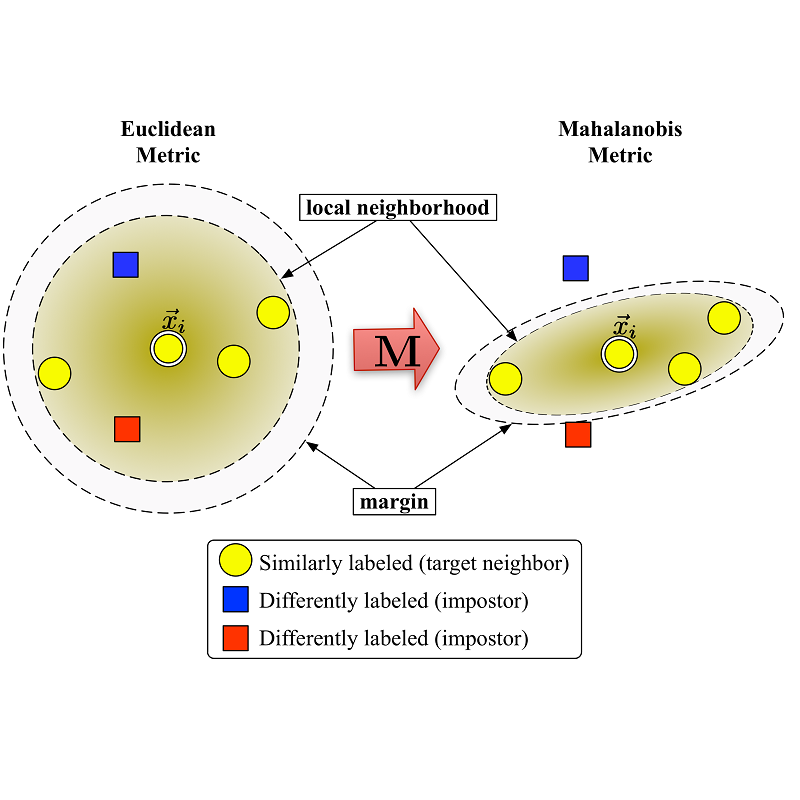
Language model pre-training (LMPT) has achieved remarkable results in natural language understanding. However, LMPT is much less successful in non-natural language domains like protein sequences, revealing a crucial discrepancy between the various sequential domains. Here, we posit that while LMPT can effectively model per-token relations, it fails at modeling per-sequence relations in non-natural language domains. To this end, we develop a framework that couples LMPT with deep structure-preserving metric learning to produce richer embeddings than can be obtained from LMPT alone. We examine new and existing pre-training models in this framework and theoretically analyze the framework overall. We also design experiments on a variety of synthetic datasets and new graph-augmented datasets of proteins and scientific abstracts. Our approach offers notable performance improvements on downstream tasks, including prediction of protein remote homology and classification of citation intent.
翻译:语言模型预培训在自然语言理解方面取得了显著成果,然而,在蛋白质序列等非自然语言领域,LMPT远不如在蛋白质序列等非自然语言领域成功,揭示了各个相继领域之间的重大差异。在这里,我们假设LMPT可以有效地建模亲身关系,但在非自然语言领域,它未能建模每个序列的关系。为此,我们制定了一个框架,使具有深层结构保护度的双胞胎LMPT能够产生比仅从LMPT取得的更丰富的嵌入。我们研究了这个框架中新的和现有的培训前模型,并从理论上分析了框架的总体情况。我们还设计了各种合成数据集和新的蛋白质和科学摘要图表强化数据集的实验。我们的方法为下游任务提供了显著的业绩改进,包括预测蛋白远程同质和引用意图分类。