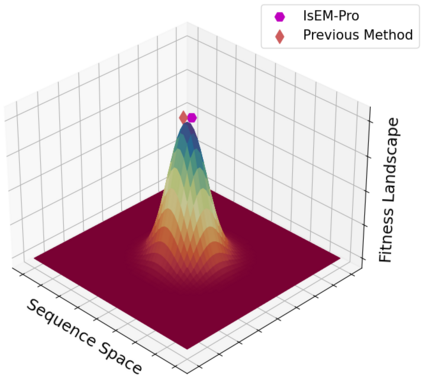
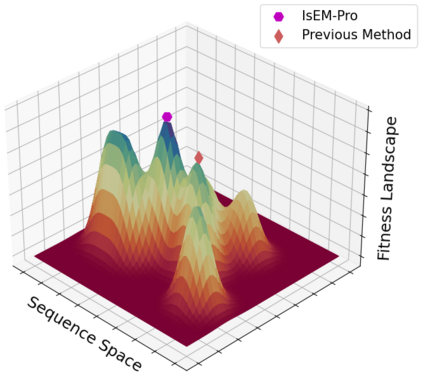
Designing protein sequences with desired biological function is crucial in biology and chemistry. Recent machine learning methods use a surrogate sequence-function model to replace the expensive wet-lab validation. How can we efficiently generate diverse and novel protein sequences with high fitness? In this paper, we propose IsEM-Pro, an approach to generate protein sequences towards a given fitness criterion. At its core, IsEM-Pro is a latent generative model, augmented by combinatorial structure features from a separately learned Markov random fields (MRFs). We develop an Monte Carlo Expectation-Maximization method (MCEM) to learn the model. During inference, sampling from its latent space enhances diversity while its MRFs features guide the exploration in high fitness regions. Experiments on eight protein sequence design tasks show that our IsEM-Pro outperforms the previous best methods by at least 55% on average fitness score and generates more diverse and novel protein sequences.
翻译:暂无翻译