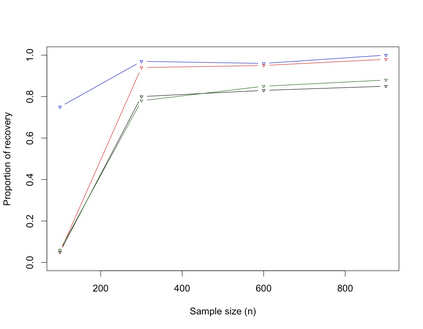
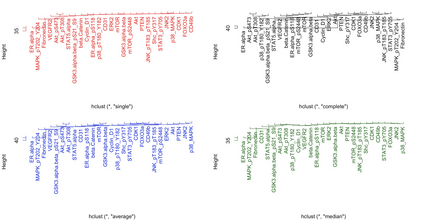
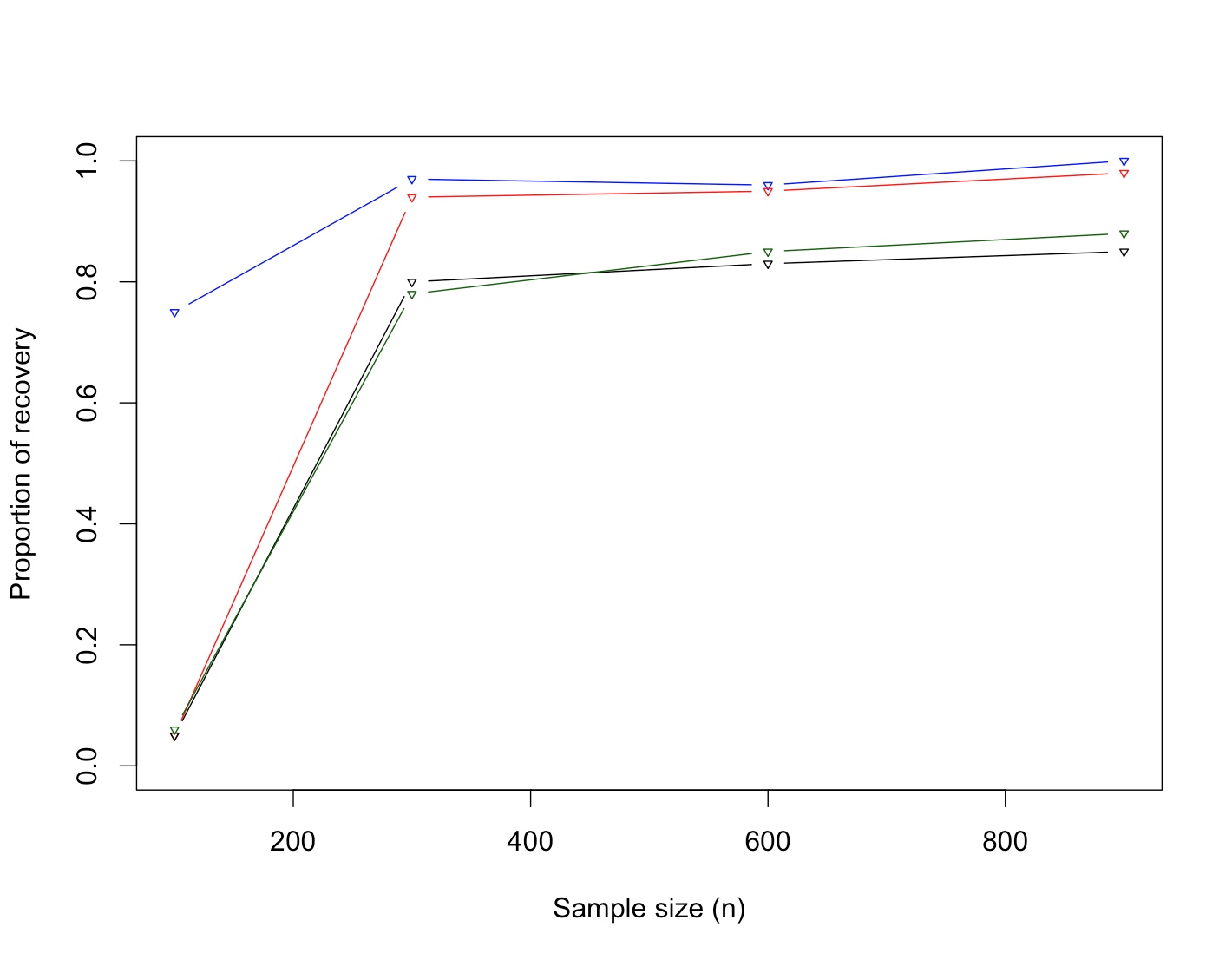
Clustering of proteins is of interest in cancer cell biology. This article proposes a hierarchical Bayesian model for protein (variable) clustering hinging on correlation structure. Starting from a multivariate normal likelihood, we enforce the clustering through prior modeling using angle based unconstrained reparameterization of correlations and assume a truncated Poisson distribution (to penalize the large number of clusters) as prior on the number of clusters. The posterior distributions of the parameters are not in explicit form and we use a reversible jump Markov chain Monte Carlo (RJMCMC) based technique is used to simulate the parameters from the posteriors. The end products of the proposed method are estimated cluster configuration of the proteins (variables) along with the number of clusters. The Bayesian method is flexible enough to cluster the proteins as well as the estimate the number of clusters. The performance of the proposed method has been substantiated with extensive simulation studies and one protein expression data with a hereditary disposition in breast cancer where the proteins are coming from different pathways.
翻译:蛋白质组群与癌症细胞生物学有关。 本文建议采用一种高层次的贝叶西亚蛋白( 变式) 组合模型, 将蛋白质( 变式) 组群放在相关结构上。 从多变的正常可能性开始, 我们通过使用基于角度的、 不加限制的对相关关系进行重新计数的先建模强制集聚, 并假设比先前的组群数多得多的Poisson分布( 以惩罚大量组群) 。 参数的后表分布没有明确的形式, 我们使用一种基于可逆跳跃 Markov 链 Monte Carlo ( RJMCMC) 的技术来模拟子宫的参数 。 拟议方法的最终产品是蛋白质( 变式) 和组群集数的估计组合组合组合。 贝伊斯 方法足够灵活地将蛋白质组群群群群聚集起来, 以及估计组群集的数量。 拟议方法的性表现得到了广泛的模拟研究的证实, 一个蛋白质表达数据得到了来自不同途径的乳腺癌遗传性处理。