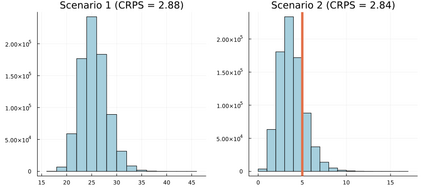
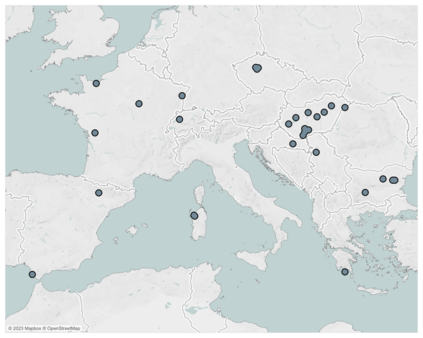
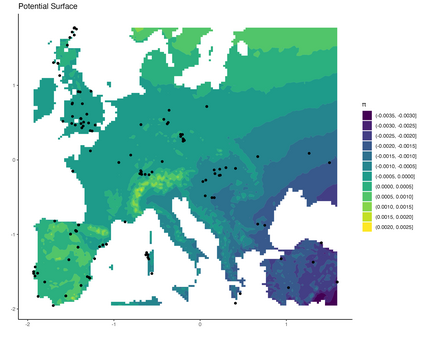
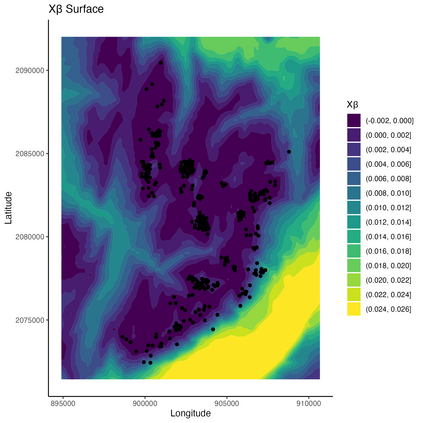
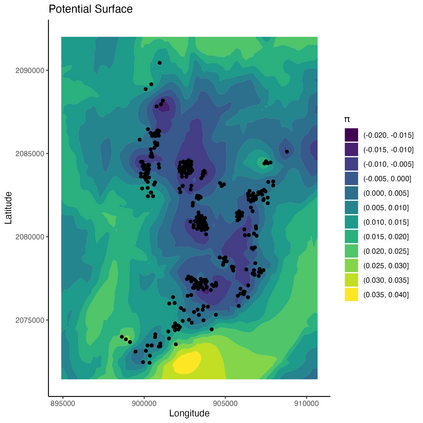
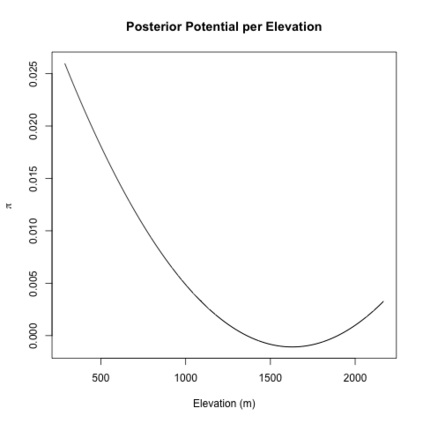
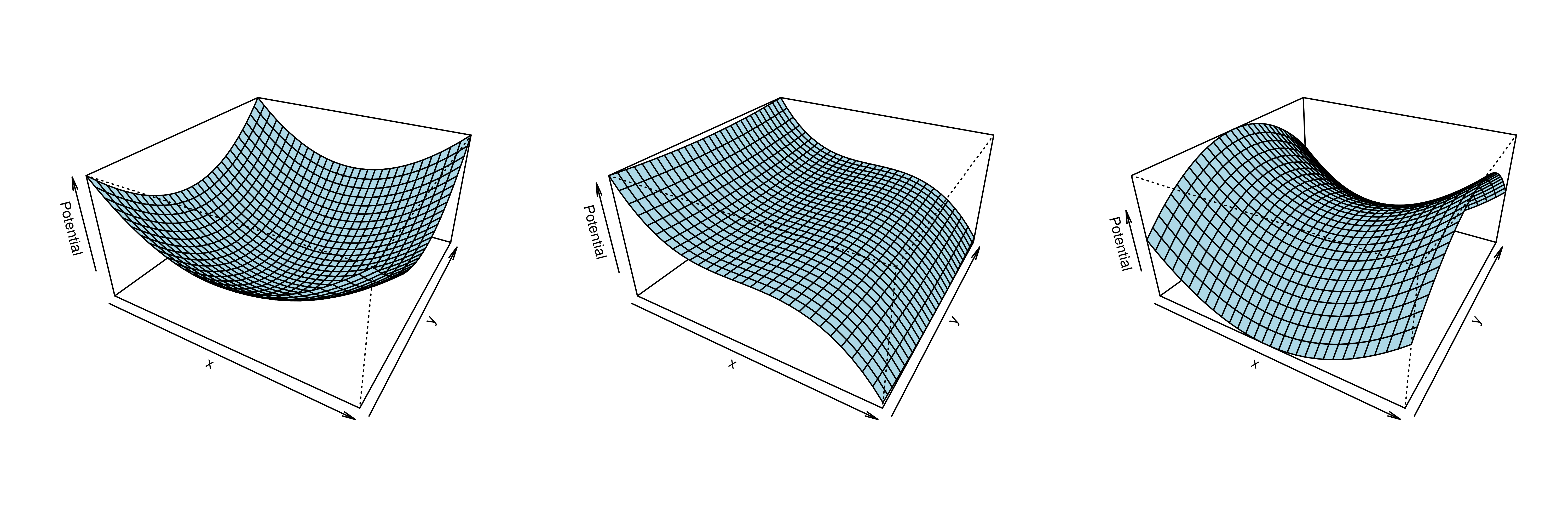
Mechanistic statistical models are commonly used to study the flow of biological processes. For example, in landscape genetics, the aim is to infer mechanisms that govern gene flow in populations. Existing statistical approaches in landscape genetics do not account for temporal dependence in the data and may be computationally prohibitive. We infer mechanisms with a Bayesian hierarchical dyadic model that scales well with large data sets and that accounts for spatial and temporal dependence. We construct a fully-connected network comprising spatio-temporal data for the dyadic model and use normalized composite likelihoods to account for the dependence structure in space and time. Our motivation for developing a dyadic model was to account for physical mechanisms commonly found in physical-statistical models. However, a numerical solver is not required in our approach because we model first-order changes directly. We apply our methods to ancient human DNA data to infer the mechanisms that affected human movement in Bronze Age Europe.
翻译:暂无翻译