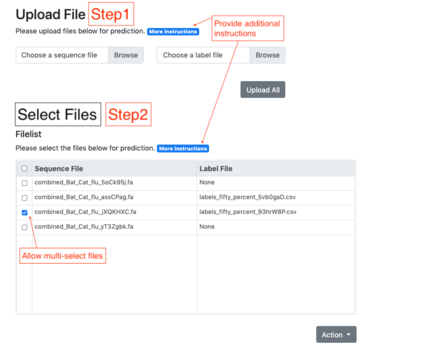
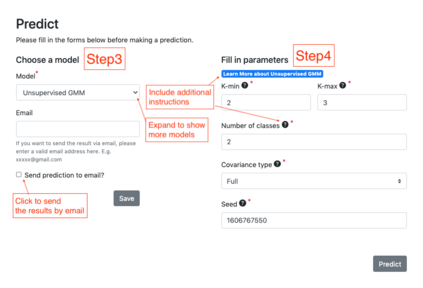
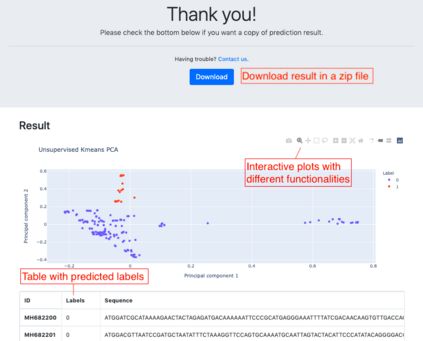
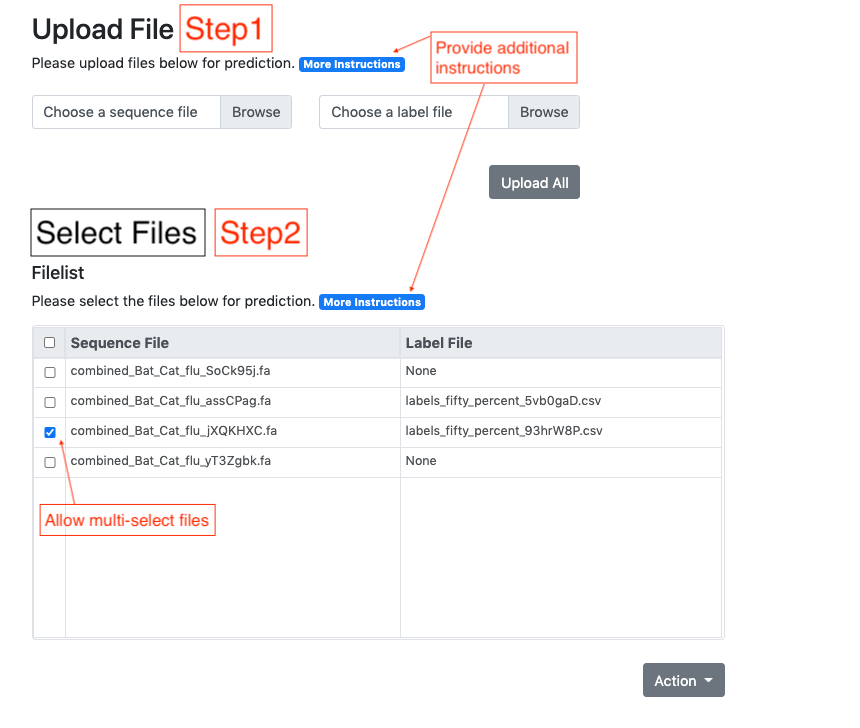
Summary: Accurate phenotype prediction from genomic sequences is a highly coveted task in biological and medical research. While machine-learning holds the key to accurate prediction in a variety of fields, the complexity of biological data can render many methodologies inapplicable. We introduce BioKlustering, a user-friendly open-source and publicly available web app for unsupervised and semi-supervised learning specialized for cases when sequence alignment and/or experimental phenotyping of all classes are not possible. Among its main advantages, BioKlustering 1) allows for maximally imbalanced settings of partially observed labels including cases when only one class is observed, which is currently prohibited in most semi-supervised methods, 2) takes unaligned sequences as input and thus, allows learning for widely diverse sequences (impossible to align) such as virus and bacteria, 3) is easy to use for anyone with little or no programming expertise, and 4) works well with small sample sizes. %This section should summarize the purpose/novel features of the program in one or two sentences. Availability and Implementation: BioKlustering (https://bioklustering.wid.wisc.edu) is a freely available web app implemented with Django, a Python-based framework, with all major browsers supported. The web app does not need any installation, and it is publicly available and open-source (https://github.com/solislemuslab/bioklustering).
翻译:摘要:基因组序列中精密的线性类型预测是生物学和医学研究中一项高度令人羡慕的任务。虽然机器学习是准确预测各个领域的关键,但生物数据的复杂性可能使许多方法不适用。我们引入了BioKlustering,这是一个方便用户的开放源码,并公开提供网络应用程序,用于所有类别的序列对齐和(或)实验性口味无法做到的不监督和半监督的学习。它的主要优点之一是,BioKlustering 1)允许在只观察到一个类的情况下,包括只看到一个案例时,部分观察到的标签设置极不平衡。目前大多数半监督方法都禁止这样做,但生物数据的复杂性使许多方法无法适用。我们引入了BioKlustering,因此,可以学习诸如病毒和细菌等广泛多样的序列(可能统一),3)对于没有多少或没有方案编制专门知识的任何人来说很容易使用,以及4)与小型样本大小一起工作。% 本节应在一或两句话中总结程序的目的/鼻子特征。 版本和执行:BioKluskwas brustering (http://dlivestowaster) a slifliflifliflivesto) a suplifusto