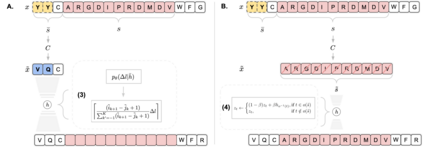
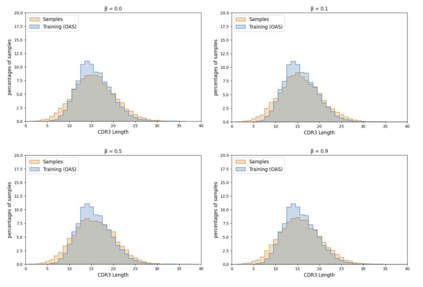
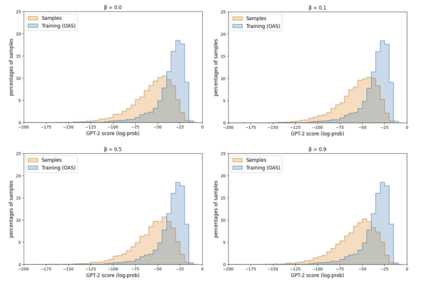
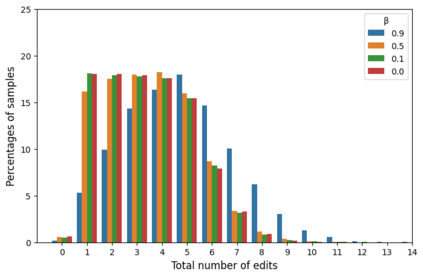
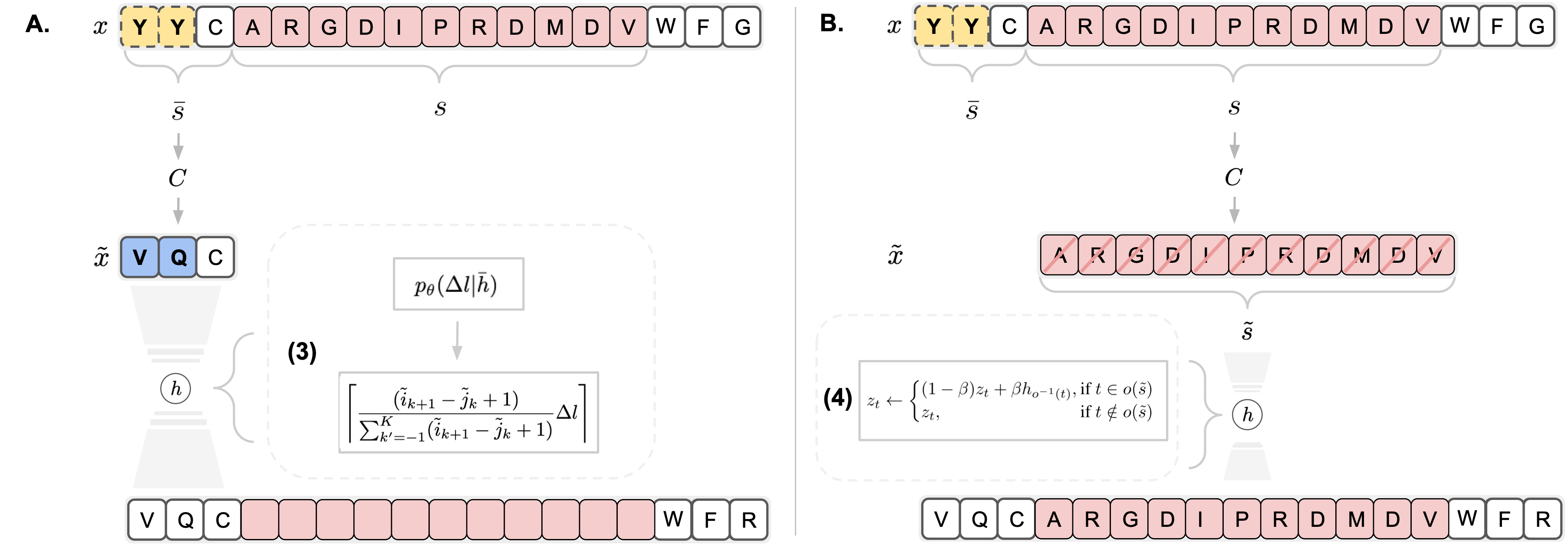
Deep generative modeling for biological sequences presents a unique challenge in reconciling the bias-variance trade-off between explicit biological insight and model flexibility. The deep manifold sampler was recently proposed as a means to iteratively sample variable-length protein sequences by exploiting the gradients from a function predictor. We introduce an alternative approach to this guided sampling procedure, multi-segment preserving sampling, that enables the direct inclusion of domain-specific knowledge by designating preserved and non-preserved segments along the input sequence, thereby restricting variation to only select regions. We present its effectiveness in the context of antibody design by training two models: a deep manifold sampler and a GPT-2 language model on nearly six million heavy chain sequences annotated with the IGHV1-18 gene. During sampling, we restrict variation to only the complementarity-determining region 3 (CDR3) of the input. We obtain log probability scores from a GPT-2 model for each sampled CDR3 and demonstrate that multi-segment preserving sampling generates reasonable designs while maintaining the desired, preserved regions.
翻译:生物序列的深层基因模型在调和明确生物洞察力和模型灵活性之间的偏差取舍方面提出了独特的挑战。最近提出了深层多元取样器,作为利用功能预测器的梯度反复采样可变长蛋白序列的手段。我们为这种有指导的采样程序引入了另一种办法,即多层保留采样,以便能够在输入序列中指定保留和未保存的区段,从而直接纳入具体领域的知识,从而将差异限制在特定区域。我们通过培训两种模型来介绍抗体设计的有效性:深层多采样器和GPT-2语言模型,涉及近600万个重链序列,并附有IGHV1-18基因。在采样时,我们只将差异限制在投入的互补-确定区域3 (CDR3) 上,我们从每个取样的CDR3 的GPT-2 模型中获得逻辑分数,并表明多层保存采样能产生合理的设计,同时维持理想的保存区域。