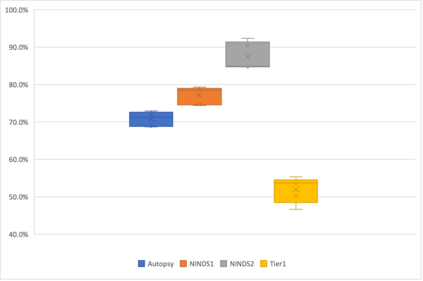
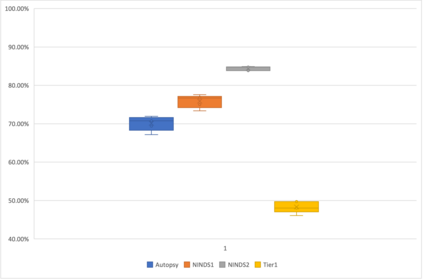
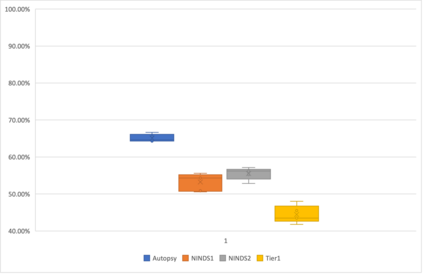
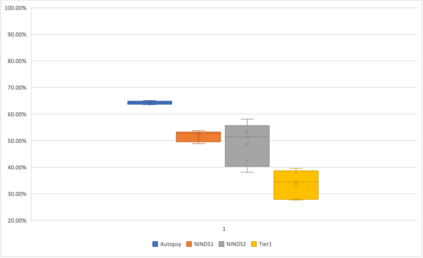
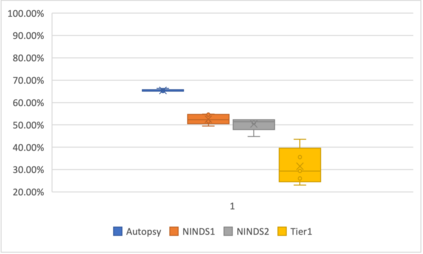
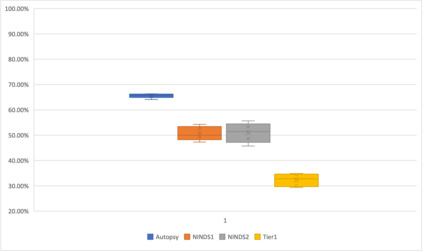
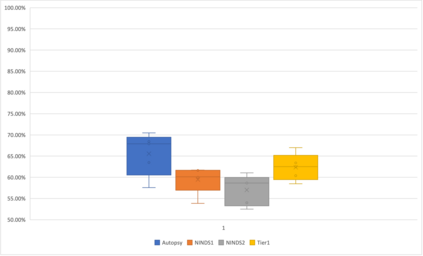
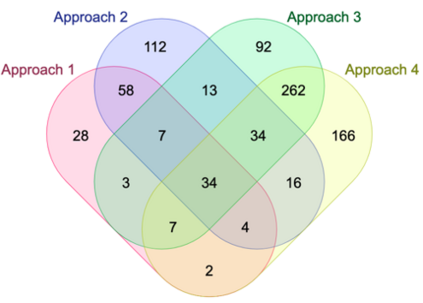
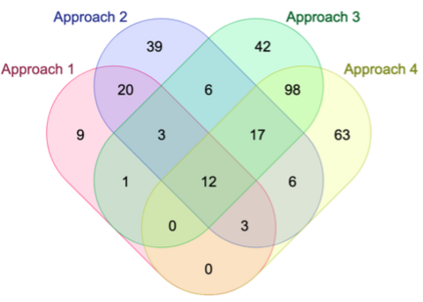
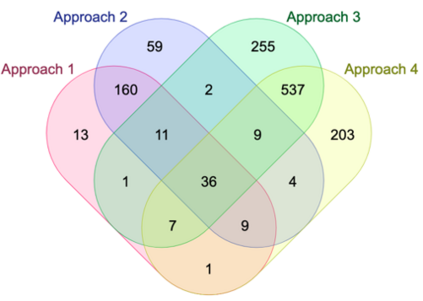
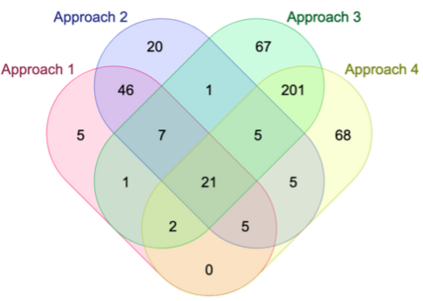
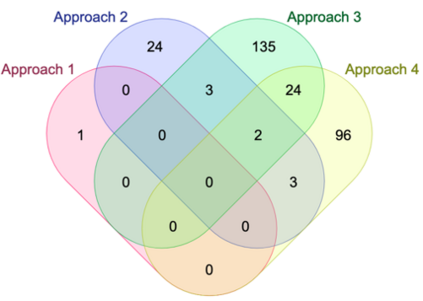
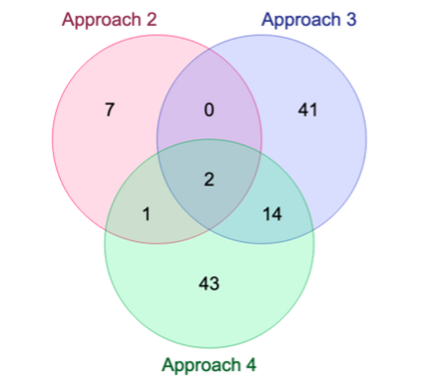
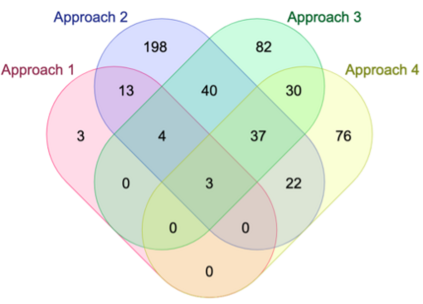
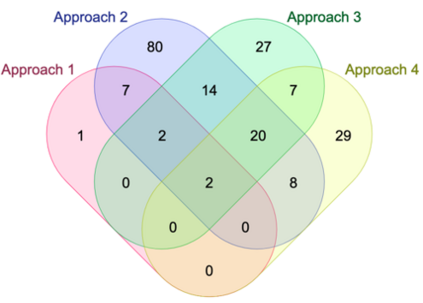
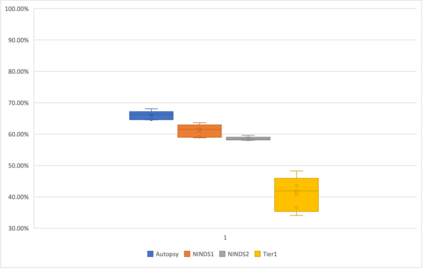
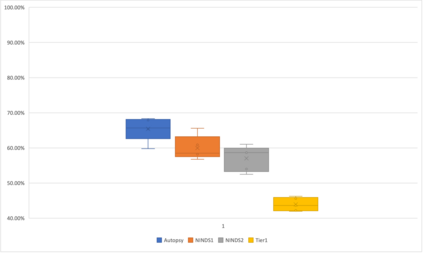
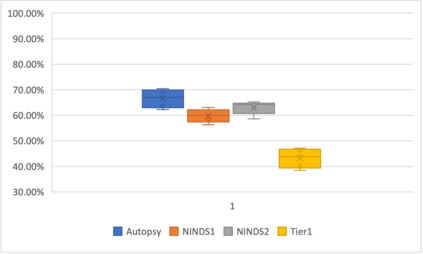
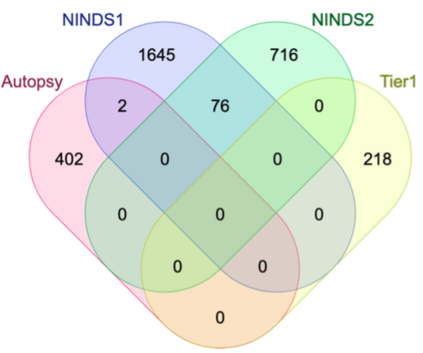
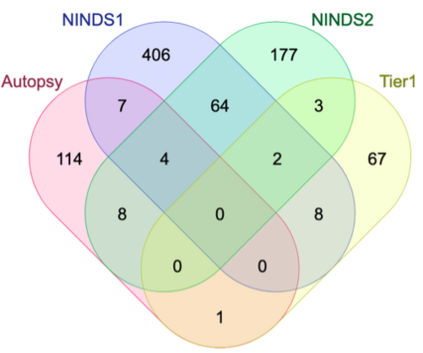
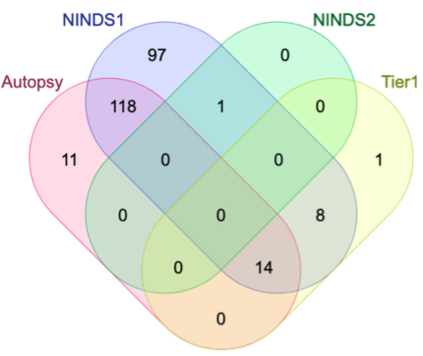
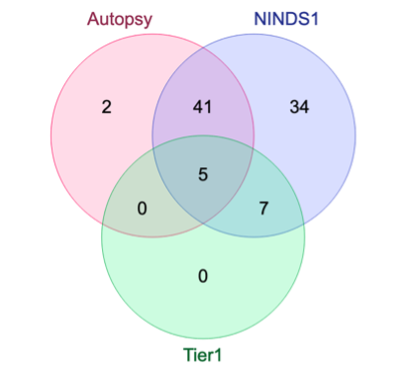
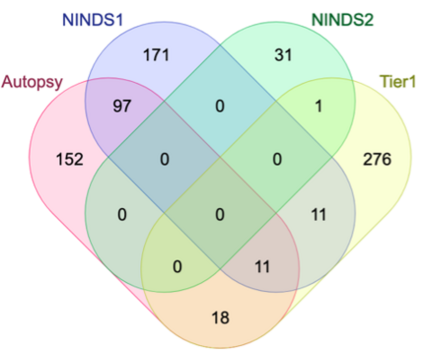
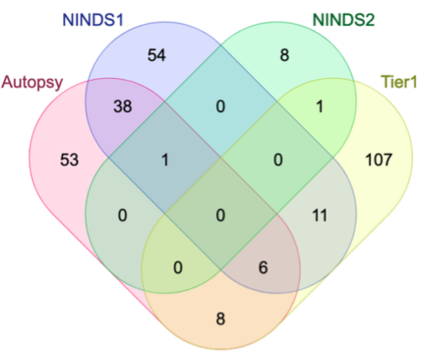
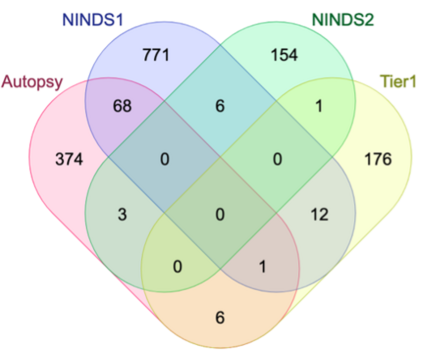
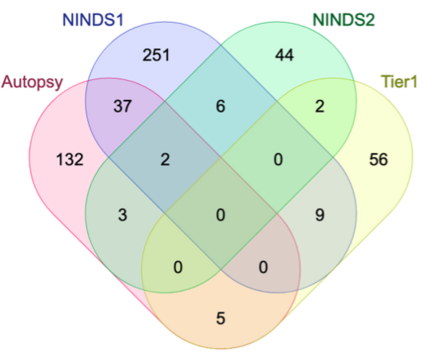
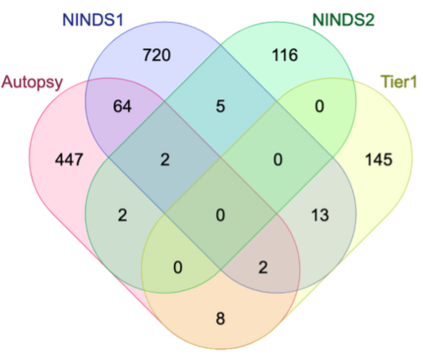
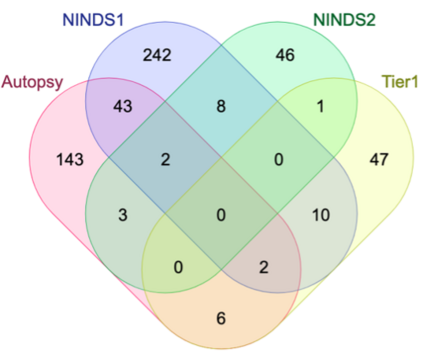
Genome-Wide Association Studies (GWAS) help identify genetic variations in people with diseases such as Parkinson's disease (PD), which are less common in those without the disease. Thus, GWAS data can be used to identify genetic variations associated with the disease. Feature selection and machine learning approaches can be used to analyze GWAS data and identify potential disease biomarkers. However, GWAS studies have technical variations that affect the reproducibility of identified biomarkers, such as differences in genotyping platforms and selection criteria for individuals to be genotyped. To address this issue, we collected five GWAS datasets from the database of Genotypes and Phenotypes (dbGaP) and explored several data integration strategies. We evaluated the agreement among different strategies in terms of the Single Nucleotide Polymorphisms (SNPs) that were identified as potential PD biomarkers. Our results showed a low concordance of biomarkers discovered using different datasets or integration strategies. However, we identified fifty SNPs that were identified at least twice, which could potentially serve as novel PD biomarkers. These SNPs are indirectly linked to PD in the literature but have not been directly associated with PD before. These findings open up new potential avenues of investigation.
翻译:基因组关联研究(GWAS)有助于确定患有帕金森病(PD)等疾病的人群中与不患有该疾病的人群不同的遗传变异。因此,GWAS数据可用于识别与疾病相关的遗传变异。特征选择和机器学习方法可以用于分析GWAS数据并识别潜在的疾病生物标志物。但是,GWAS研究存在技术差异,影响已确定的生物标志物的可重复性,例如不同的基因分型平台和选择要进行基因分型的个体的标准。为解决这个问题,我们收集了来自数据库of Genotypes and Phenotypes (dbGaP)的五个GWAS数据集,并探索了几种数据整合策略。我们评估了不同策略之间在被识别为潜在PD生物标志物的单核苷酸多态性(SNP)方面的一致性。我们的结果显示,使用不同数据集或整合策略识别出的生物标志物一致性较低。但是,我们已确定至少出现两次的五十个SNP,这可能作为新的PD生物标志物。这些SNP与PD间接相关,但以前没有直接与PD相关联。这些发现开启了新的潜在研究领域。