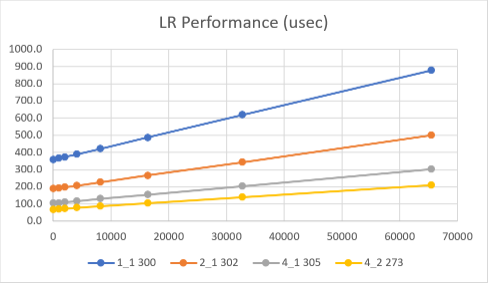
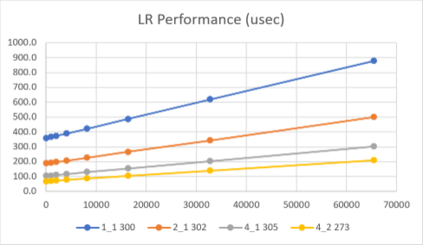
Molecular Dynamics (MD) simulations play a central role in physics-driven drug discovery. MD applications often use the Particle Mesh Ewald (PME) algorithm to accelerate electrostatic force computations, but efficient parallelization has proven difficult due to the high communication requirements of distributed 3D FFTs. In this paper, we present the design and implementation of a scalable PME algorithm that runs on a cluster of Intel Stratix 10 FPGAs and can handle FFT sizes appropriate to address real-world drug discovery projects (grids up to $128^3$). To our knowledge, this is the first work to fully integrate all aspects of the PME algorithm (charge spreading, 3D FFT/IFFT, and force interpolation) within a distributed FPGA framework. The design is fully implemented with OpenCL for flexibility and ease of development and uses 100 Gbps links for direct FPGA-to-FPGA communications without the need for host interaction. We present experimental data up to 4 FPGAs (e.g., 206 microseconds per timestep for a 65536 atom simulation and $64^3$ 3D FFT), outperforming GPUs. Additionally, we discuss design scalability on clusters with differing topologies up to 64 FPGAs (with expected performance greater than all known GPU implementations) and integration with other hardware components to form a complete molecular dynamics application. We predict best-case performance of 6.6 microseconds per timestep on 64 FPGAs.
翻译:分子动态模拟(MD)在物理驱动的药物发现中发挥着核心作用。MD应用程序经常使用Parte Mesh Ewald(PME)算法加速静电力计算,但由于3DFFT的分布式3DFFT的通信要求很高,高效平行化证明是困难的。在本文中,我们介绍在Intel Stratix 10 FPGAs 10 FPGAs的一个集群上运行的可缩放的PME算法的设计和实施,并可以处理适合处理真实世界毒品发现项目的FFFFT规模(Grigs最多达128 3美元)。据我们了解,这是将PME算法(3D FFT/IFF和武力内插)的所有方面完全纳入一个分布式的FPGGGGGGA框架框架框架框架范围内的通信要求很高。设计与OCLIL一起全面实施灵活和方便开发的PGGGGGGGGA-FGA通信,不需要进行主机式互动。我们所知道的4 FFGGGGGGA-FA系统内部动态应用中的所有实验数据最多可达65536,比OGFAFA的软化应用。