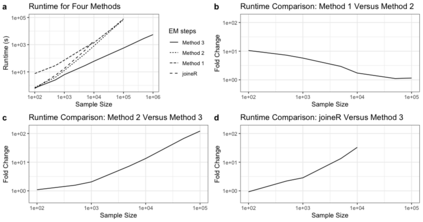
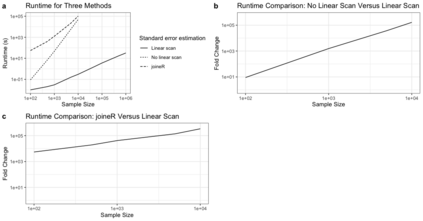
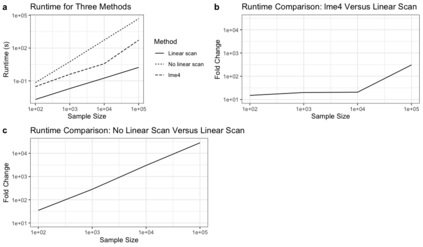
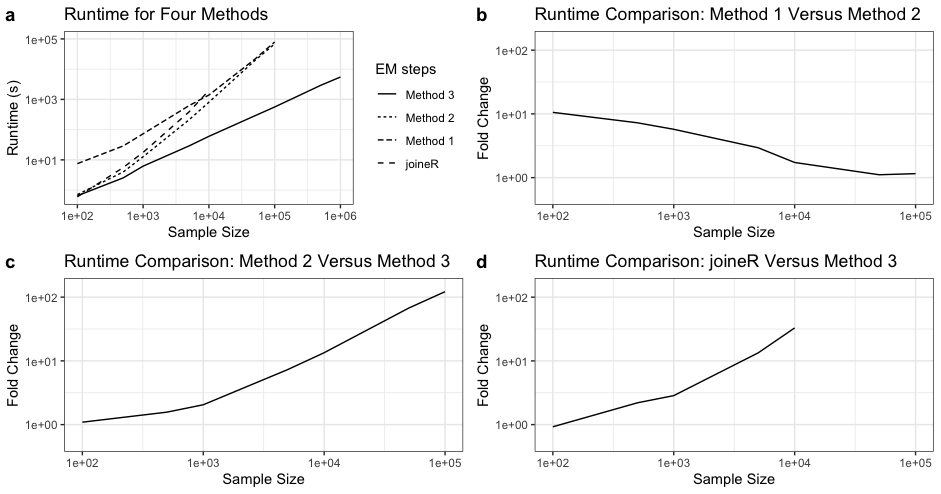
Semiparametric joint models of longitudinal and competing risks data are computationally costly and their current implementations do not scale well to massive biobank data. This paper identifies and addresses some key computational barriers in a semiparametric joint model for longitudinal and competing risks survival data. By developing and implementing customized linear scan algorithms, we reduce the computational complexities from $O(n^2)$ or $O(n^3)$ to $O(n)$ in various components including numerical integration, risk set calculation, and standard error estimation, where $n$ is the number of subjects. Using both simulated and real world biobank data, we demonstrate that these linear scan algorithms generate drastic speed-up of up to hundreds of thousands fold when $n>10^4$, sometimes reducing the run-time from days to minutes. We have developed an R-package, FastJM, based on the proposed algorithms for joint modeling of longitudinal and time-to-event data with and without competing risks, and made it publicly available on the Comprehensive R Archive Network (CRAN) at \url{https://CRAN.R-project.org/package=FastJM}.
翻译:纵向和相互竞争的风险数据的半参数联合模型计算成本很高,而目前实施的这些模型对大型生物库数据的影响并不大。本文件在纵向和相互竞争的风险生存数据的半参数联合模型中确定并解决了一些关键的计算障碍。我们通过开发和实施定制的线性扫描算法,将各种组成部分的计算复杂性从O(n)2美元或O(n)3美元降低到O(n)美元,其中包括数字整合、风险计算和标准错误估计,其中以美元为主题。我们利用模拟和真实的世界生物库数据,证明这些线性扫描算法在10-4美元时产生了高达数十万倍的急剧速度,有时将运行时间从几天缩短到几分钟。我们根据拟议的长度和时间-时间-时间-活动数据的联合模型算法开发了一个R包件“快速”和“快速”软件,在风险和没有相互竞争的情况下将其公布在\url{http://CRAN.R-projro.org/package=FARMY}的综合档案网络(CRANAN)上。