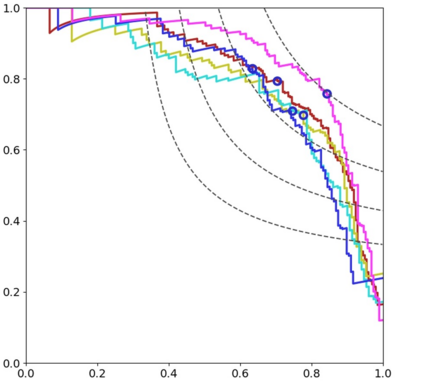
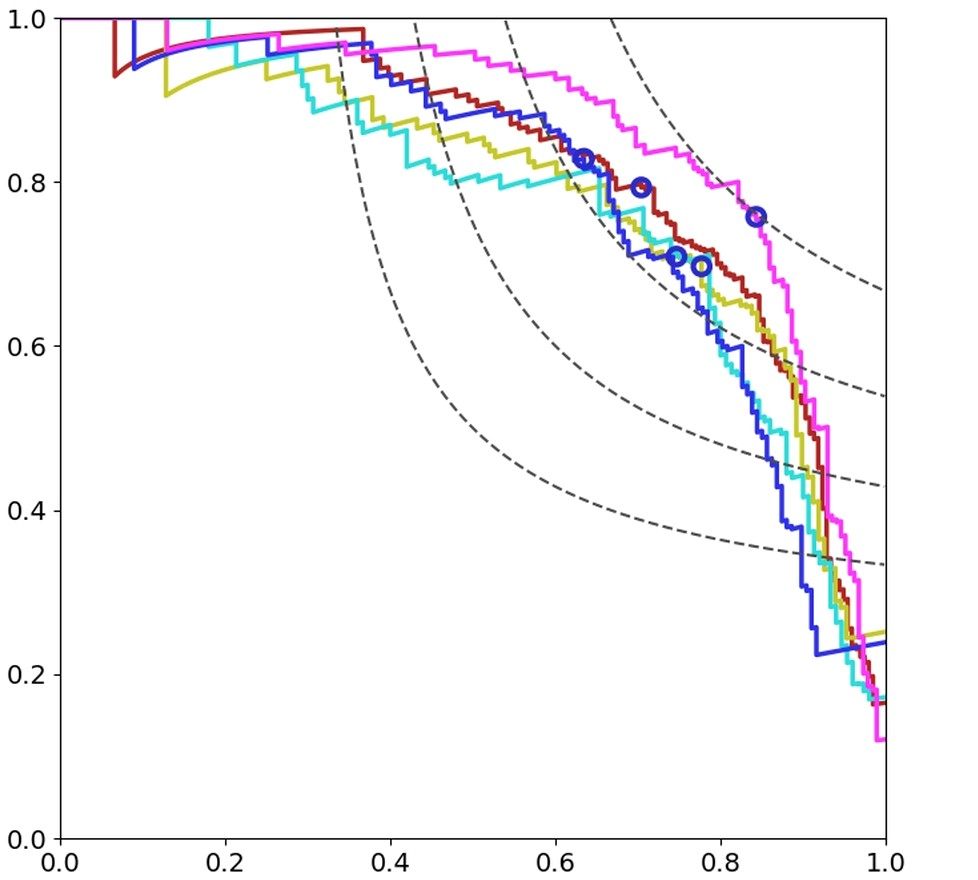
Automated detection of mitotic figures in histopathology images is a challenging task: here, we present the different steps that describe the strategy we applied to participate in the MIDOG 2021 competition. The purpose of the competition was to evaluate the generalization of solutions to images acquired with unseen target scanners (hidden for the participants) under the constraint of using training data from a limited set of four independent source scanners. Given this goal and constraints, we joined the challenge by proposing a straight-forward solution based on a combination of state-of-the-art deep learning methods with the aim of yielding robustness to possible scanner-related distributional shifts at inference time. Our solution combines methods that were previously shown to be efficient for mitosis detection: hard negative mining, extensive data augmentation, rotation-invariant convolutional networks. We trained five models with different splits of the provided dataset. The subsequent classifiers produced F1-scores with a mean and standard deviation of 0.747+/-0.032 on the test splits. The resulting ensemble constitutes our candidate algorithm: its automated evaluation on the preliminary test set of the challenge returned a F1-score of 0.6828.
翻译:在组织病理学图象中自动检测光学图象是一个具有挑战性的任务:在这里,我们提出不同的步骤,说明我们参与MIDOG 2021 竞争的战略,竞争的目的是评价对利用秘密目标扫描仪(参与者的隐蔽)获得的图像解决方案的概括性,这些图像的解决方案是在使用一组有限的四个独立源扫描仪的培训数据的限制下取得的(参与者的隐蔽的)。鉴于这个目标和制约因素,我们加入挑战,提出一个直向前进的解决方案,其基础是结合最先进的深层次学习方法,目的是在推断时使扫描仪可能相关的分布变化具有稳健性。我们的解决办法结合了以前证明对分解检测有效的方法:硬式负式采矿、广泛的数据增强、旋转-变异共振网络。我们训练了五种模型,对所提供的数据集进行了不同的分割。随后的分类方法在测试分解上产生了F1核心,其平均和标准偏差为0.747+/0.032。由此产生的元素构成我们的候选人算法:对F828号初步测试返回的挑战集的F8进行自动评价。
相关内容
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem