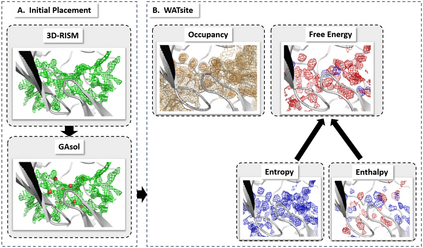
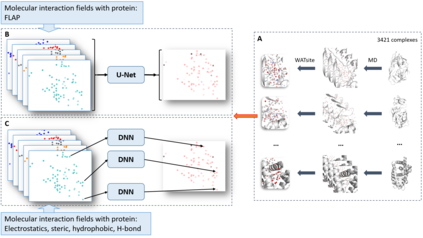
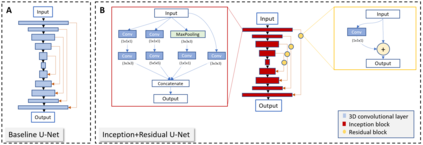
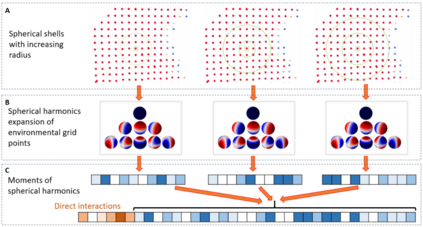
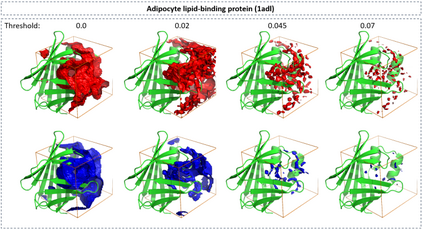
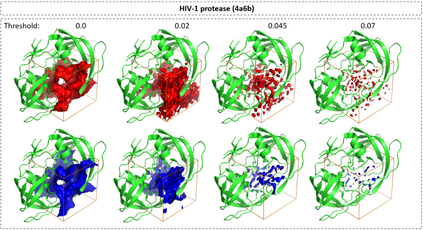
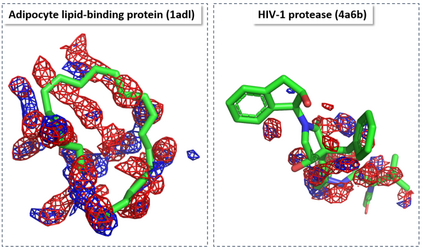
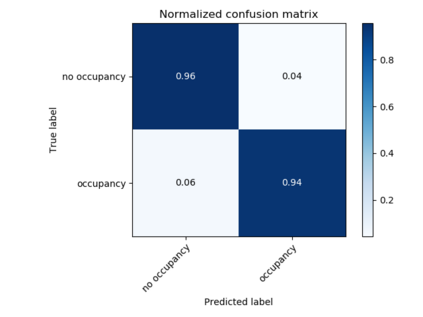
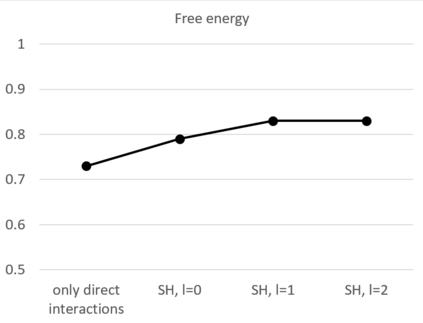
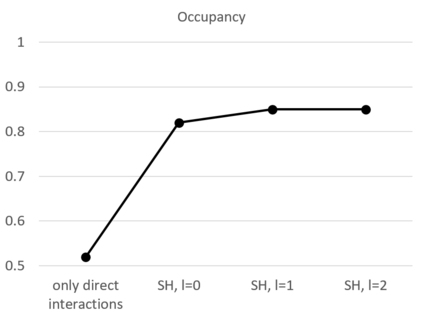
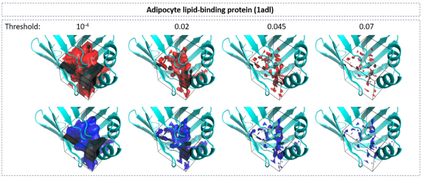
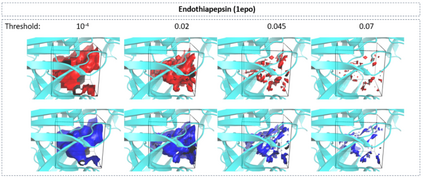
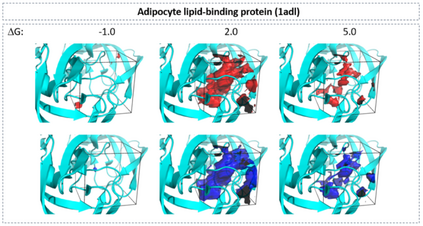
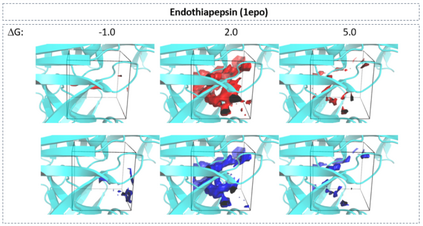
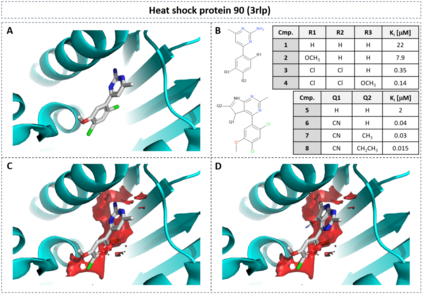
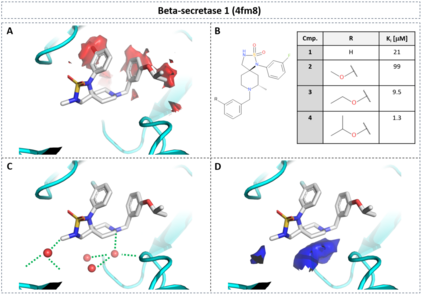
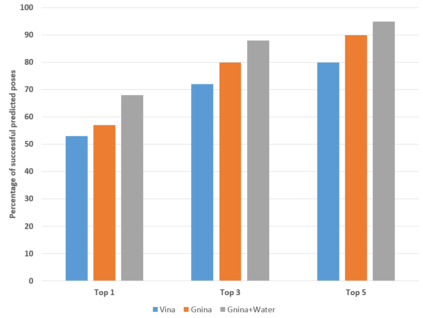
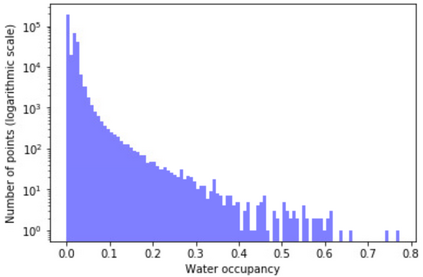
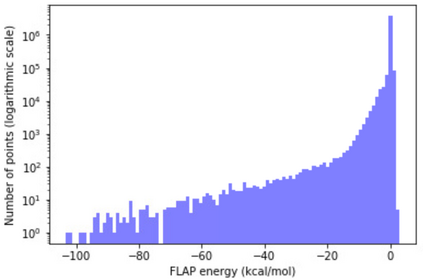
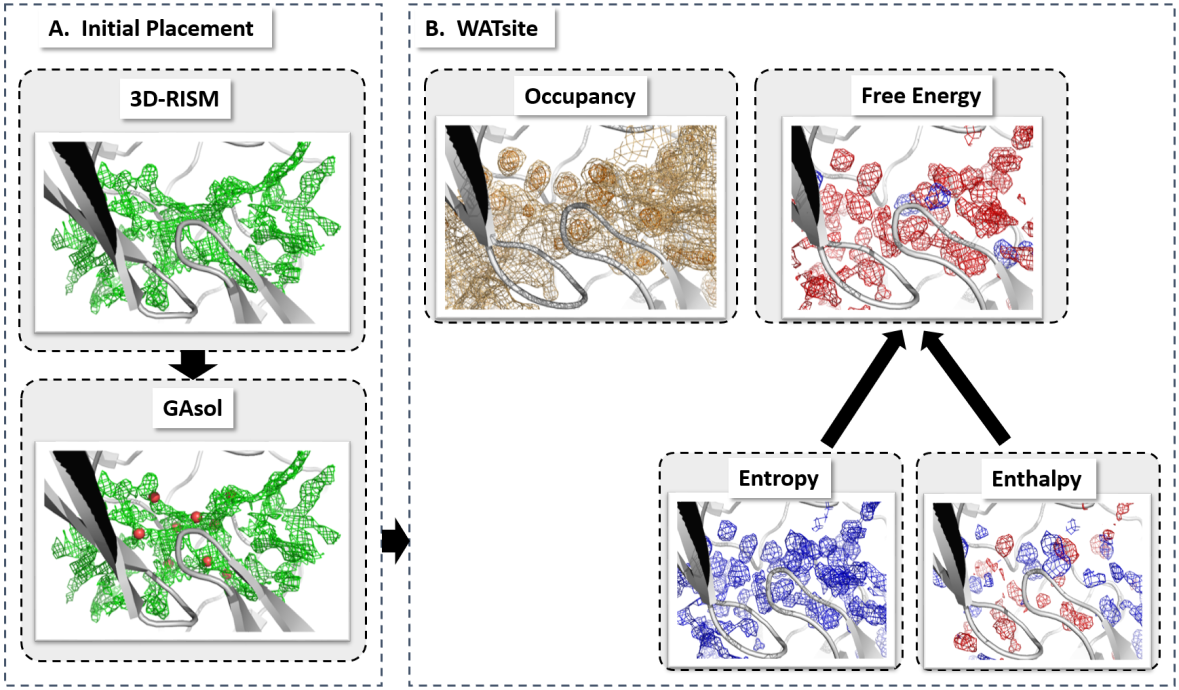
The calculation of thermodynamic properties of biochemical systems typically requires the use of resource-intensive molecular simulation methods. One example thereof is the thermodynamic profiling of hydration sites, i.e. high-probability locations for water molecules on the protein surface, which play an essential role in protein-ligand associations and must therefore be incorporated in the prediction of binding poses and affinities. To replace time-consuming simulations in hydration site predictions, we developed two different types of deep neural-network models aiming to predict hydration site data. In the first approach, meshed 3D images are generated representing the interactions between certain molecular probes placed on regular 3D grids, encompassing the binding pocket, with the static protein. These molecular interaction fields are mapped to the corresponding 3D image of hydration occupancy using a neural network based on an U-Net architecture. In a second approach, hydration occupancy and thermodynamics were predicted point-wise using a neural network based on fully-connected layers. In addition to direct protein interaction fields, the environment of each grid point was represented using moments of a spherical harmonics expansion of the interaction properties of nearby grid points. Application to structure-activity relationship analysis and protein-ligand pose scoring demonstrates the utility of the predicted hydration information.
翻译:生物化学系统的热力特性的计算通常需要使用资源密集的分子模拟方法,其中一个例子是对水合点的热力分析,即蛋白质表面的水分子高概率位置,在蛋白质-离心和关联中起着重要作用,因此必须纳入对装质和亲近性的预测中。为了取代水合点预测中耗时的模拟,我们开发了两种不同类型的深度神经网络模型,旨在预测水合点数据。在第一个方法中,生成了3D型网格图像,代表了固定的3D电网中某些分子探测器与静态蛋白质的相互作用。这些分子互动字段是用来绘制在基于U-Net结构的神经网络中对应的3D水合体图像的。在第二个方法中,水合和热力反应是利用基于完全连接层的神经网络预测。除了直接的蛋白互动场外,每个电联点的环境都代表着固定式3D网格网格内装口袋的固定分子探测器与静态蛋白质的相互作用。这些分子互动域域域域与基于U-网络结构结构的预测性电压特性分析,显示电压结构的电压特性。