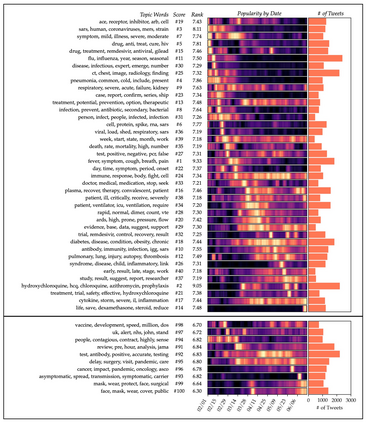
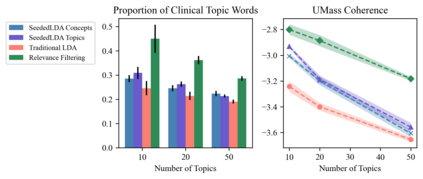
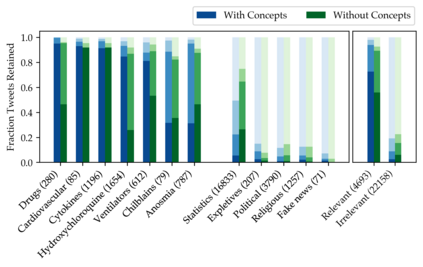
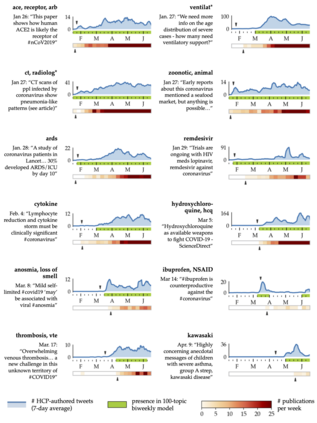
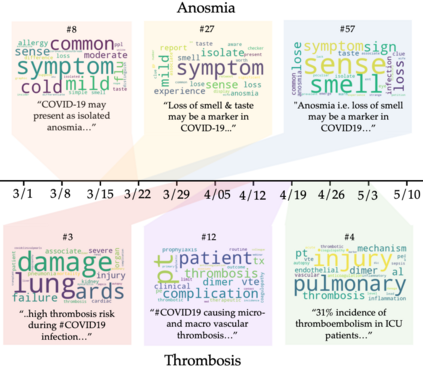
The rapid evolution of the COVID-19 pandemic has underscored the need to quickly disseminate the latest clinical knowledge during a public-health emergency. One surprisingly effective platform for healthcare professionals (HCPs) to share knowledge and experiences from the front lines has been social media (for example, the "#medtwitter" community on Twitter). However, identifying clinically-relevant content in social media without manual labeling is a challenge because of the sheer volume of irrelevant data. We present an unsupervised, iterative approach to mine clinically relevant information from social media data, which begins by heuristically filtering for HCP-authored texts and incorporates topic modeling and concept extraction with MetaMap. This approach identifies granular topics and tweets with high clinical relevance from a set of about 52 million COVID-19-related tweets from January to mid-June 2020. We also show that because the technique does not require manual labeling, it can be used to identify emerging topics on a week-to-week basis. Our method can aid in future public-health emergencies by facilitating knowledge transfer among healthcare workers in a rapidly-changing information environment, and by providing an efficient and unsupervised way of highlighting potential areas for clinical research.
翻译:在公共卫生紧急情况下,COVID-19大流行病的迅速演变凸显了迅速传播最新临床知识的必要性,在公共卫生紧急情况下,保健专业人员分享第一线知识和经验的一个令人惊讶的有效平台是社交媒体(例如推特上的“#medTwitter”社群);然而,在社会媒体中发现临床相关内容而不用人工标签是一种挑战,因为相关数据数量庞大,因此不需要人工标签。我们从社交媒体数据中对与地雷临床相关的信息采用了一种不受监督的迭接方法,从黑过滤HCP所撰写的文本开始,并结合MetaMap进行主题建模和概念提取。这一方法确定了从2020年1月至6月中旬的一组约5 200万个COVID-19相关推特中具有高度临床相关性的颗粒议题和推特。我们还表明,由于技术不需要人工标签,因此可以使用周至周内新出现的主题。我们的方法可以帮助未来的公共卫生紧急情况,方法是在快速变化的信息环境中促进保健工作者之间的知识转让,并为临床研究领域提供高效和未经监督的潜在领域。