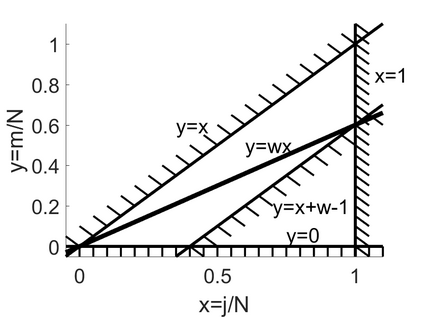
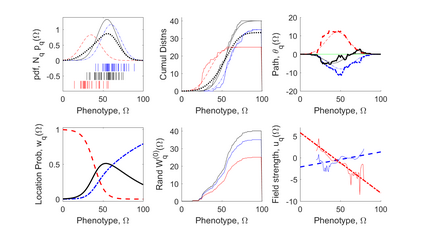
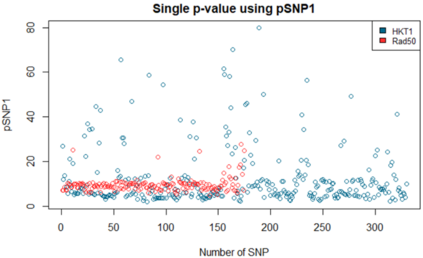
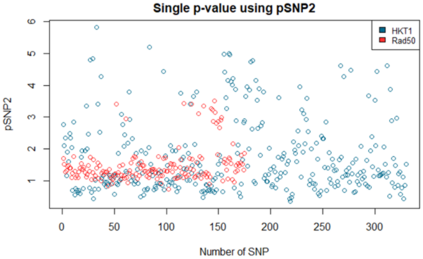
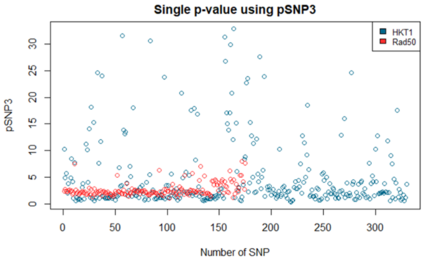
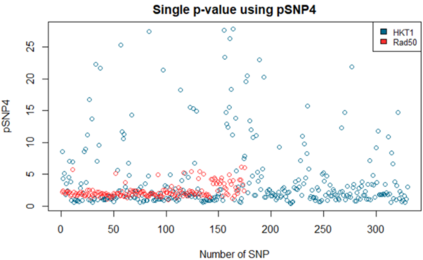
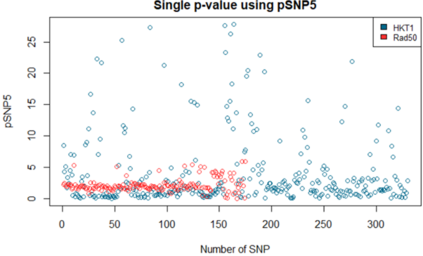
We show how field- and information theory can be used to quantify the relationship between genotype and phenotype in cases where phenotype is a continuous variable. Given a sample population of phenotype measurements, from various known genotypes, we show how the ordering of phenotype data can lead to quantification of the effect of genotype. This method does not assume that the data has a Gaussian distribution, it is particularly effective at extracting weak and unusual dependencies of genotype on phenotype. However, in cases where data has a special form, (eg Gaussian), we observe that the effective phenotype field has a special form. We use asymptotic analysis to solve both the forward and reverse formulations of the problem. We show how $p$-values can be calculated so that the significance of correlation between phenotype and genotype can be quantified. This provides a significant generalisation of the traditional methods used in genome-wide association studies GWAS. We derive a field-strength which can be used to deduce how the correlations between genotype and phenotype, and their impact on the distribution of phenotypes.
翻译:我们展示了如何在苯型是一个连续变量的情况下使用字段和信息理论来量化基因类型与苯型之间的关系。考虑到从各种已知基因型中对苯型测量的样本群,我们展示了对苯型数据的排序如何导致对基因型效应的量化。这种方法不假定数据具有高斯分布,对于提取在苯型上对基因型的脆弱和异常依赖性特别有效。然而,在数据具有特殊形式的情况下(如高山),我们观察到有效的苯型字段有特殊形式。我们使用随机分析来解决问题的前向和反向配方。我们展示了如何计算美元值,以便量化苯型与基因型之间关联的重要性。这为基因组-全协会研究GWAS使用的传统方法提供了一种显著的概括性。我们得出了一种外向力,可以用来推断基因型和苯型类型对基因型分布的影响和苯型的关联性。我们展示了一种外向力,可以用来推断基因型类型和苯型类型对基因型分布的关联性。