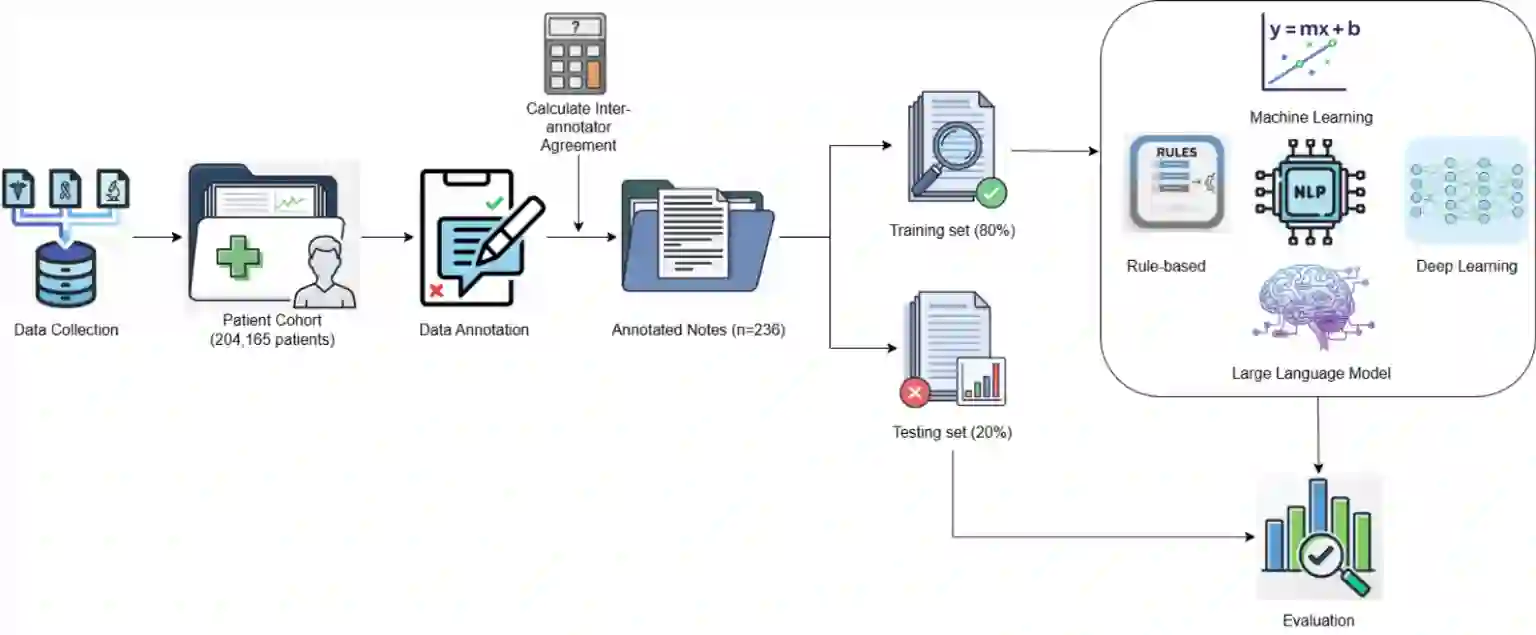
Objective: Fluoropyrimidines are widely prescribed for colorectal and breast cancers, but are associated with toxicities such as hand-foot syndrome and cardiotoxicity. Since toxicity documentation is often embedded in clinical notes, we aimed to develop and evaluate natural language processing (NLP) methods to extract treatment and toxicity information. Materials and Methods: We constructed a gold-standard dataset of 236 clinical notes from 204,165 adult oncology patients. Domain experts annotated categories related to treatment regimens and toxicities. We developed rule-based, machine learning-based (Random Forest, Support Vector Machine [SVM], Logistic Regression [LR]), deep learning-based (BERT, ClinicalBERT), and large language models (LLM)-based NLP approaches (zero-shot and error-analysis prompting). Models used an 80:20 train-test split. Results: Sufficient data existed to train and evaluate 5 annotated categories. Error-analysis prompting achieved optimal precision, recall, and F1 scores (F1=1.000) for treatment and toxicities extraction, whereas zero-shot prompting reached F1=1.000 for treatment and F1=0.876 for toxicities extraction.LR and SVM ranked second for toxicities (F1=0.937). Deep learning underperformed, with BERT (F1=0.873 treatment; F1= 0.839 toxicities) and ClinicalBERT (F1=0.873 treatment; F1 = 0.886 toxicities). Rule-based methods served as our baseline with F1 scores of 0.857 in treatment and 0.858 in toxicities. Discussion: LMM-based approaches outperformed all others, followed by machine learning methods. Machine and deep learning approaches were limited by small training data and showed limited generalizability, particularly for rare categories. Conclusion: LLM-based NLP most effectively extracted fluoropyrimidine treatment and toxicity information from clinical notes, and has strong potential to support oncology research and pharmacovigilance.
翻译:目的:氟尿嘧啶类药物广泛用于结直肠癌和乳腺癌的治疗,但常伴随手足综合征和心脏毒性等不良反应。由于毒性记录常嵌入临床笔记中,本研究旨在开发并评估自然语言处理(NLP)方法,以自动提取治疗方案及毒性信息。材料与方法:我们从204,165名成年肿瘤患者的临床笔记中构建了包含236份笔记的金标准数据集。领域专家对治疗方案和毒性相关类别进行了标注。我们开发了基于规则、基于机器学习(随机森林、支持向量机[SVM]、逻辑回归[LR])、基于深度学习(BERT、ClinicalBERT)以及基于大语言模型(LLM)的NLP方法(包括零样本提示和错误分析提示)。模型采用80:20的训练集-测试集划分。结果:有足够数据用于训练和评估5个标注类别。错误分析提示在治疗方案和毒性提取方面取得了最优的精确率、召回率和F1分数(F1=1.000),而零样本提示在治疗方案提取上达到F1=1.000,在毒性提取上达到F1=0.876。逻辑回归和支持向量机在毒性提取上表现次优(F1=0.937)。深度学习方法表现欠佳,BERT(治疗方案F1=0.873;毒性F1=0.839)和ClinicalBERT(治疗方案F1=0.873;毒性F1=0.886)。基于规则的方法作为基线,在治疗方案和毒性提取上的F1分数分别为0.857和0.858。讨论:基于LLM的方法优于其他所有方法,其次是机器学习方法。机器学习和深度学习方法的性能受限于训练数据量小,泛化能力有限,尤其对于罕见类别。结论:基于LLM的NLP方法能最有效地从临床笔记中提取氟尿嘧啶治疗方案及毒性信息,在肿瘤学研究和药物警戒领域具有巨大应用潜力。