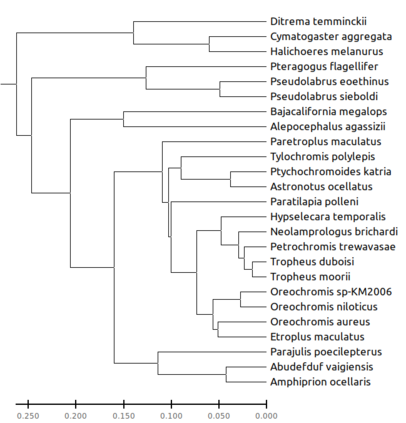
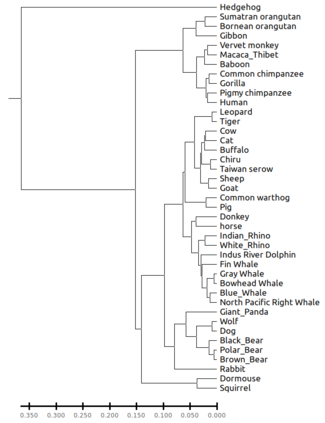
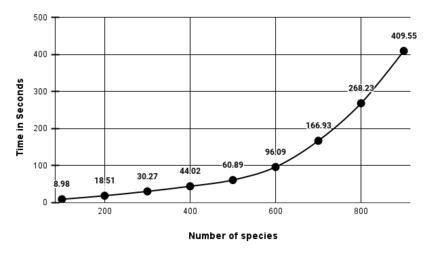
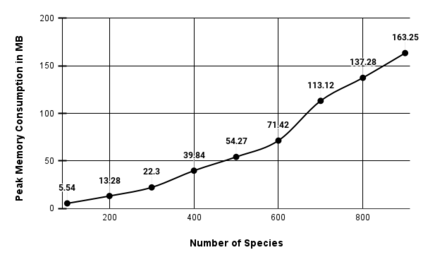
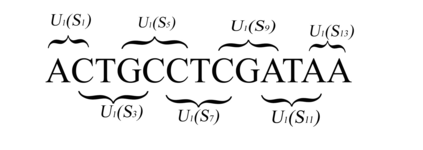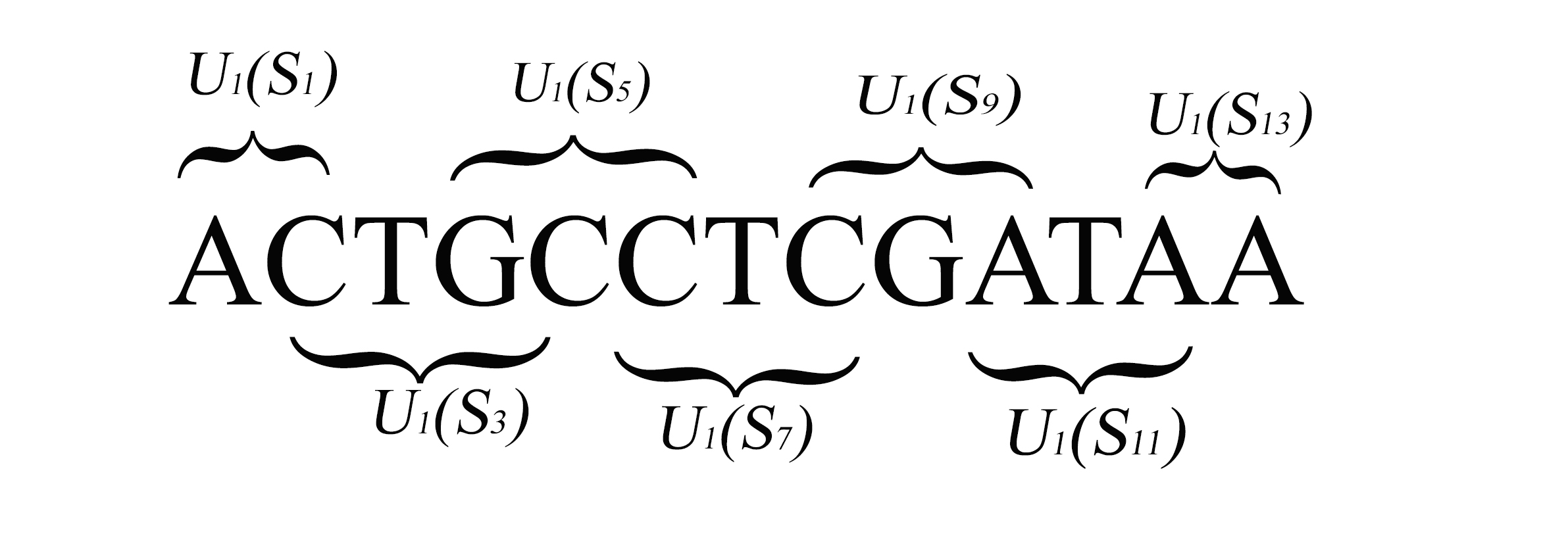We construct a compact vector representation on $\mathbb{R}^{24}$ of a DNA sequence of arbitrary length. Each component of this vector is obtained from a representative sequence, the elements of which are the values realized by a function $\Gamma$. The function $\Gamma$, so defined, acts on neighborhoods of arbitrary radius that are located at strategic positions within the DNA sequence. $\Gamma$ carries complete information about the local multiplicity of the nucleotides as a consequence of the uniqueness of prime factorisation of integer. The two parameters characterizing the radius and location of the neighbourhoods are fixed by comparing the phylogenetic tree we find through our algorithm with standard results for the $\beta$ -globin gene sequences of eleven different species. Remarkably, the time complexity for this similarity analysis turns out to be $\mathcal{O}(n)$. Using the values of the two fitting parameters so obtained, the method is further applied to analyze mitochondrial genome sequences.
翻译:暂无翻译