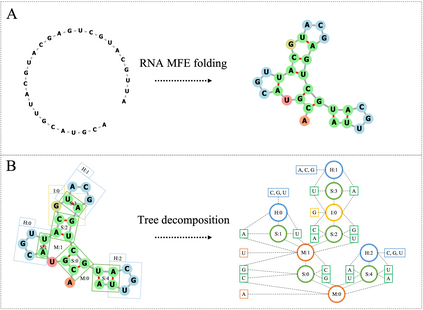
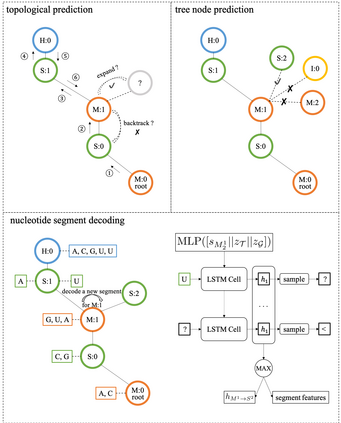
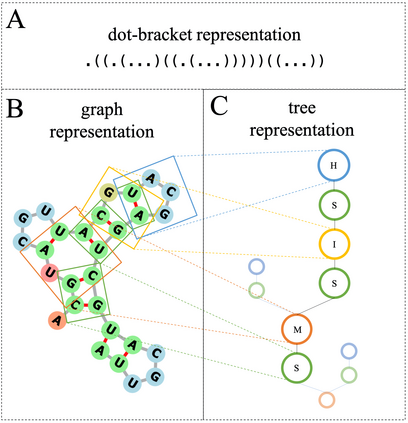
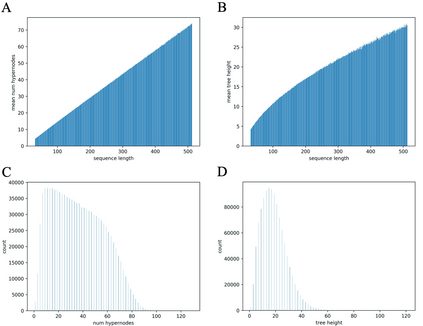
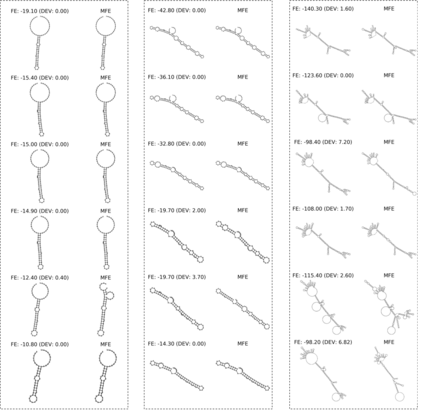
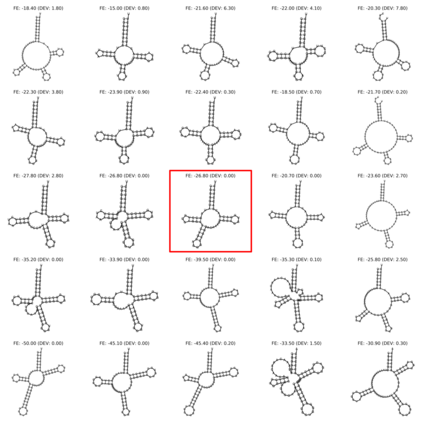
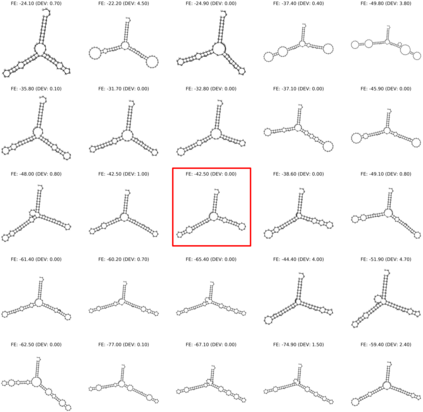
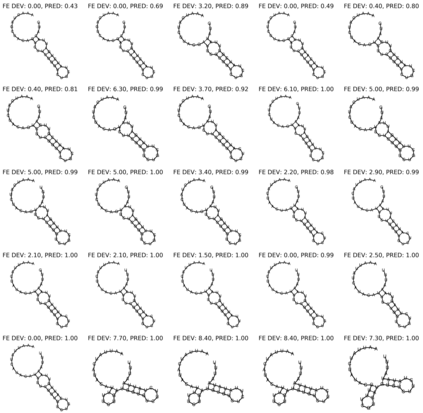
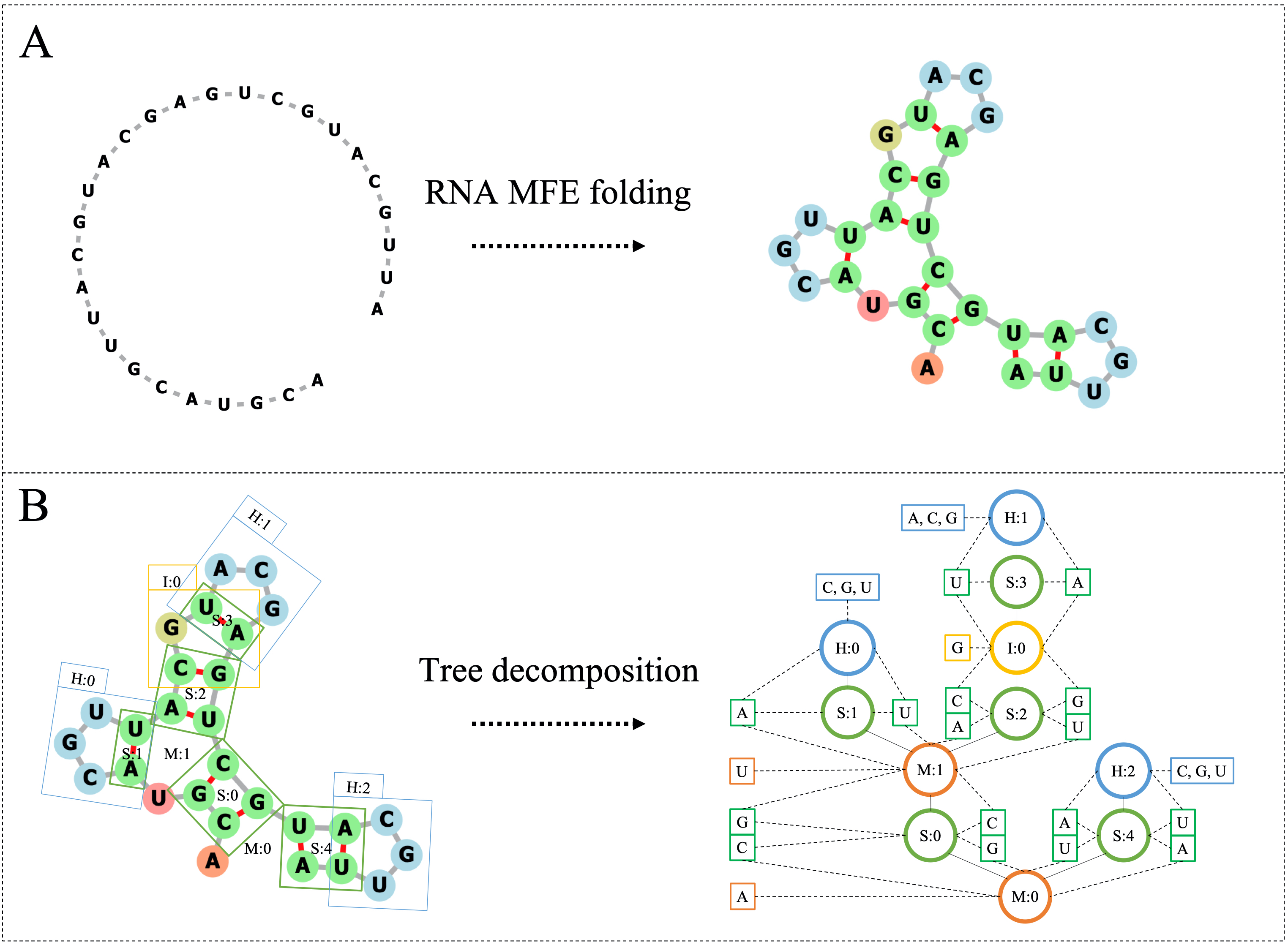
Our work is concerned with the generation and targeted design of RNA, a type of genetic macromolecule that can adopt complex structures which influence their cellular activities and functions. The design of large scale and complex biological structures spurs dedicated graph-based deep generative modeling techniques, which represents a key but underappreciated aspect of computational drug discovery. In this work, we investigate the principles behind representing and generating different RNA structural modalities, and propose a flexible framework to jointly embed and generate these molecular structures along with their sequence in a meaningful latent space. Equipped with a deep understanding of RNA molecular structures, our most sophisticated encoding and decoding methods operate on the molecular graph as well as the junction tree hierarchy, integrating strong inductive bias about RNA structural regularity and folding mechanism such that high structural validity, stability and diversity of generated RNAs are achieved. Also, we seek to adequately organize the latent space of RNA molecular embeddings with regard to the interaction with proteins, and targeted optimization is used to navigate in this latent space to search for desired novel RNA molecules.
翻译:我们的工作涉及RNA的产生和有针对性的设计,RNA是一种遗传巨分子,可以采用影响其细胞活动和功能的复杂结构。大型和复杂的生物结构的设计促使专门基于图形的深层基因模型技术,这是计算药物发现的一个关键但认识不足的方面。在这项工作中,我们调查代表并产生不同RNA结构模式的原则,并提议一个灵活的框架,以便在一个有意义的潜质空间中联合嵌入和生成这些分子结构及其序列。在对RNA分子结构的深刻理解下,我们最先进的编码和解码方法在分子图和连接树结构结构上运作,整合对RNA结构规律和折叠机制的强烈感性偏向,从而实现所产生RNA结构的高度有效性、稳定性和多样性。此外,我们还设法充分组织RNA分子与蛋白质相互作用的潜在空间,并采用有针对性的优化方法在这种潜在空间中导航所需的新式RNA分子。