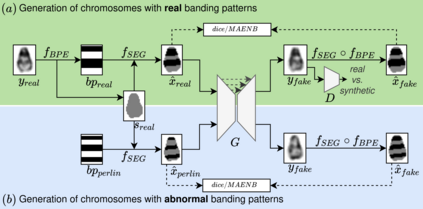
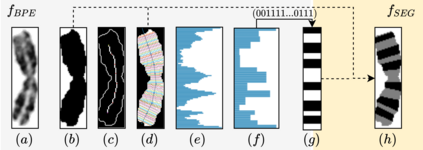
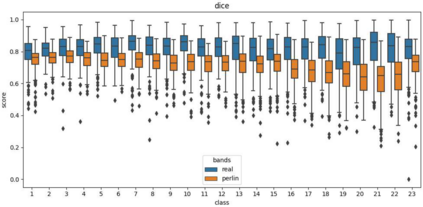
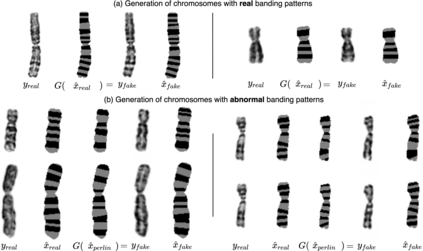
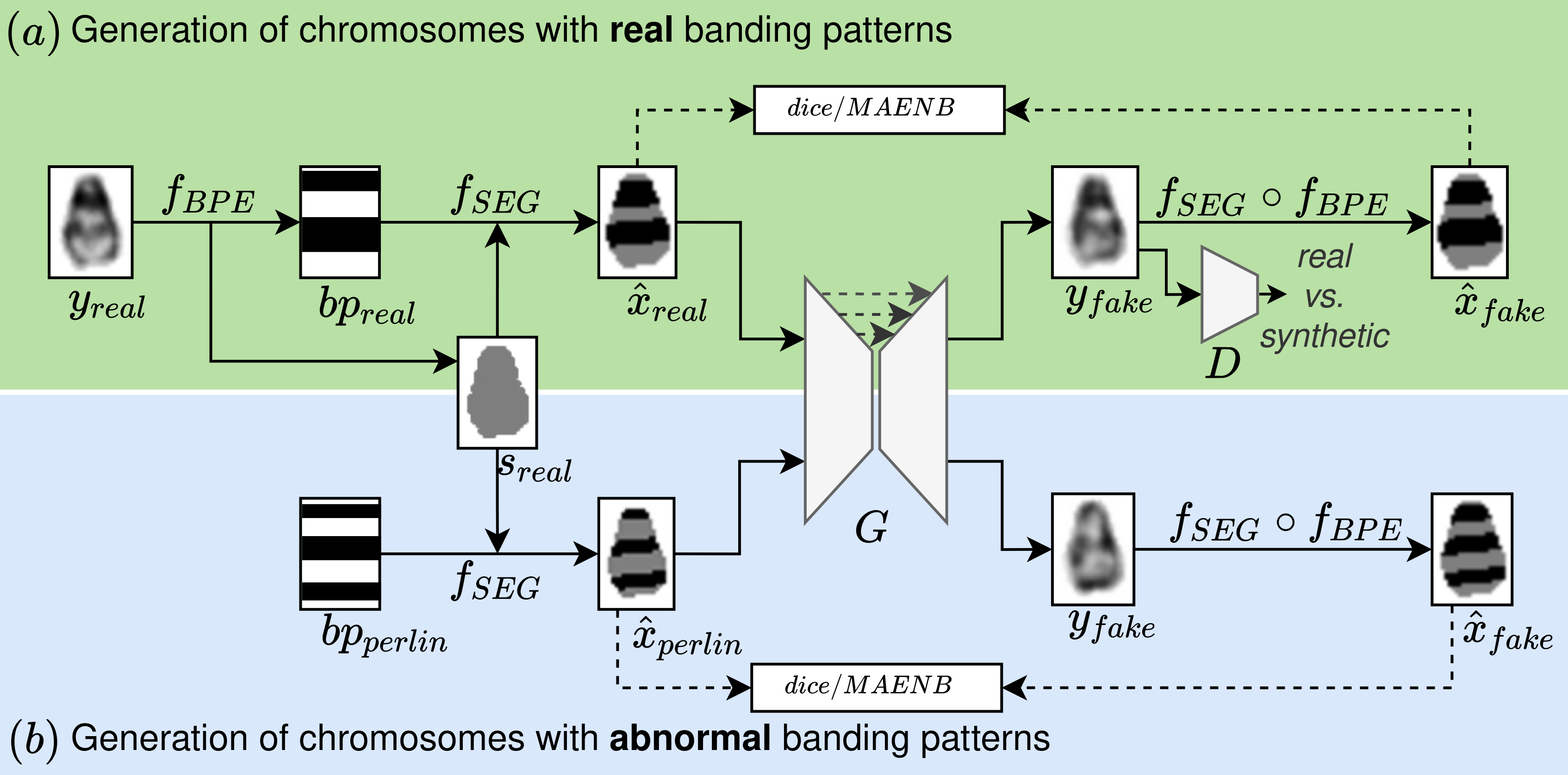
Advances in deep-learning-based pipelines have led to breakthroughs in a variety of microscopy image diagnostics. However, a sufficiently big training data set is usually difficult to obtain due to high annotation costs. In the case of banded chromosome images, the creation of big enough libraries is difficult for multiple pathologies due to the rarity of certain genetic disorders. Generative Adversarial Networks (GANs) have proven to be effective in generating synthetic images and extending training data sets. In our work, we implement a conditional adversarial network that allows generation of realistic single chromosome images following user-defined banding patterns. To this end, an image-to-image translation approach based on self-generated 2D chromosome segmentation label maps is used. Our validation shows promising results when synthesizing chromosomes with seen as well as unseen banding patterns. We believe that this approach can be exploited for data augmentation of chromosome data sets with structural abnormalities. Therefore, the proposed method could help to tackle medical image analysis problems such as data simulation, segmentation, detection, or classification in the field of cytogenetics.
翻译:深层学习管道的进步导致各种显微镜图像诊断的突破。然而,由于注释成本高,通常难以获得足够大的培训数据集。对于带带染色体图像,由于某些遗传病的罕见性,难以建立足够大的图书馆,以多种病理。基因反转网络(GANs)已证明在生成合成图像和扩大培训数据集方面是有效的。在我们的工作中,我们实施了一个有条件的对称网络,允许根据用户定义的带宽模式生成现实的单一染色体图像。为此,使用了基于自生成 2D 色谱分解标签图的图像到图像转换方法。我们的验证表明,在将染色体与看不见的带模式合成时,将产生有希望的结果。我们认为,这种方法可以用于结构异常的染色体数据集的数据增强。因此,拟议的方法可以帮助解决医学图像分析问题,如数据模拟、分解、检测或遗传学领域的数据分类。