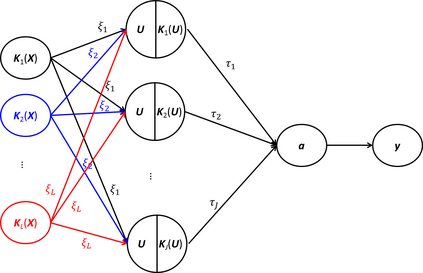
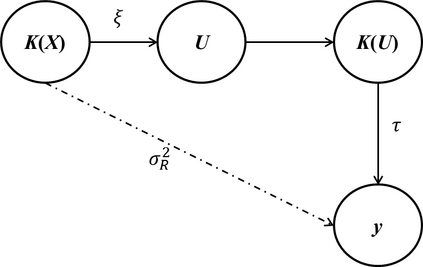
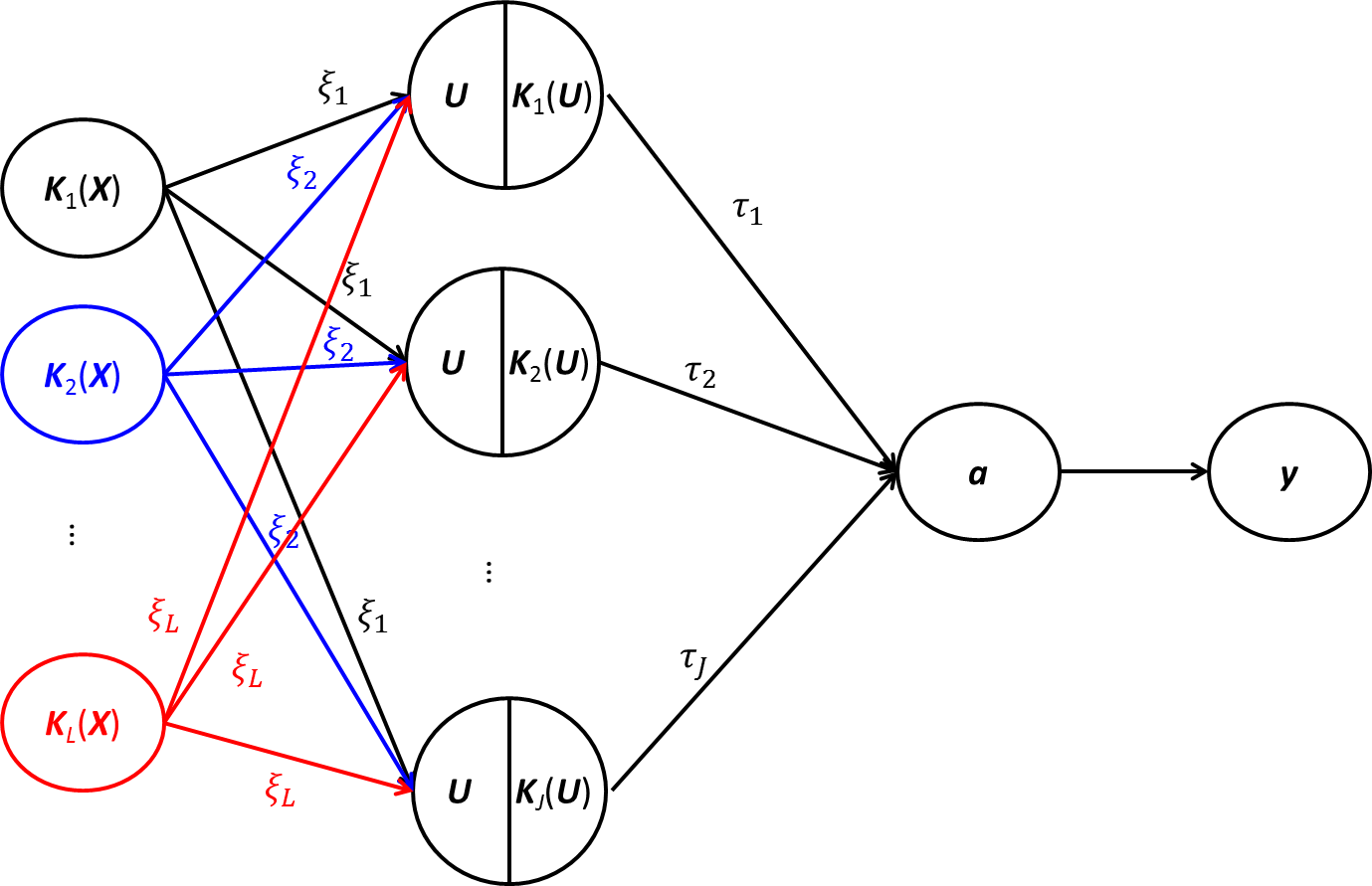
Risk prediction capitalizing on emerging human genome findings holds great promise for new prediction and prevention strategies. While the large amounts of genetic data generated from high-throughput technologies offer us a unique opportunity to study a deep catalog of genetic variants for risk prediction, the high-dimensionality of genetic data and complex relationships between genetic variants and disease outcomes bring tremendous challenges to risk prediction analysis. To address these rising challenges, we propose a kernel-based neural network (KNN) method. KNN inherits features from both linear mixed models (LMM) and classical neural networks and is designed for high-dimensional risk prediction analysis. To deal with datasets with millions of variants, KNN summarizes genetic data into kernel matrices and use the kernel matrices as inputs. Based on the kernel matrices, KNN builds a single-layer feedforward neural network, which makes it feasible to consider complex relationships between genetic variants and disease outcomes. The parameter estimation in KNN is based on MINQUE and we show, that under certain conditions, the average prediction error of KNN can be smaller than that of LMM. Simulation studies also confirm the results.
翻译:利用人类基因组新发现的风险预测为新的预测和预防战略带来了巨大的希望。尽管高通量技术产生的大量遗传数据为我们提供了一个独特的机会,可以研究关于风险预测的遗传变异物的深层目录,但遗传数据的高度性和遗传变异物与疾病结果之间的复杂关系给风险预测分析带来了巨大的挑战。为了应对这些不断上升的挑战,我们提议了一个以内核为基础的神经网络(KNN)方法。KNN继承了线性混合模型(LMM)和古典神经网络的特征,并设计了高度风险预测分析。为了处理数以百万计的变异物数据集,KNNN将遗传数据汇总到内核矩阵,并将内核矩阵用作投入。根据内核矩阵,KNN建立了一个单层进料神经网络,从而可以考虑基因变异物与疾病结果之间的复杂关系。KNNN的参数估计以MIQUE为基础,我们表明,在某些条件下,KNN的平均预测错误可能比LMM的结果要小。模拟研究也证实了结果。