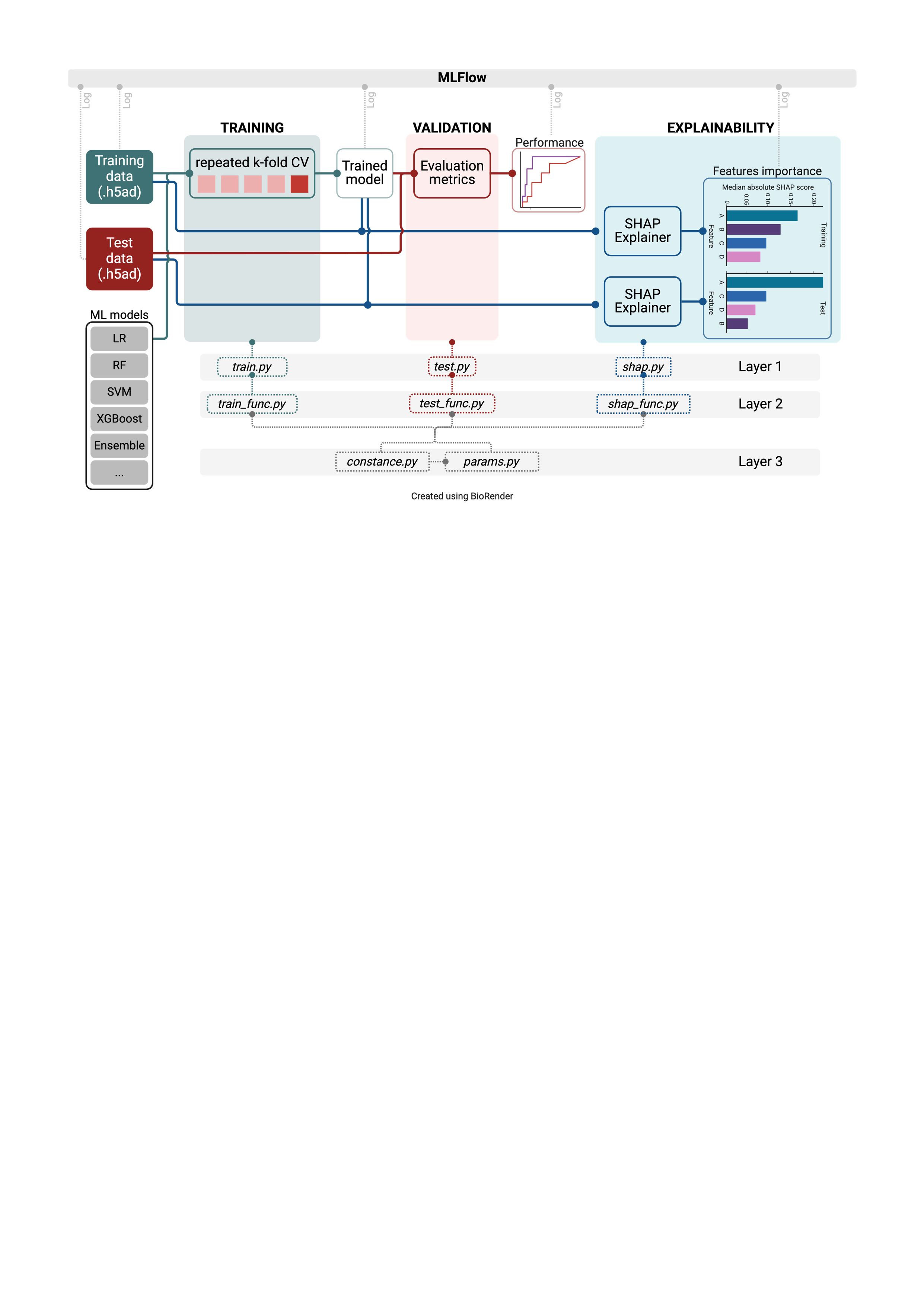
Motivation: Building and iterating machine learning models is often a resource-intensive process. In biomedical research, scientific codebases can lack scalability and are not easily transferable to work beyond what they were intended. xML-workFlow addresses this issue by providing a rapid, robust, and traceable end-to-end workflow that can be adapted to any ML project with minimal code rewriting. Results: We show a practical, end-to-end workflow that integrates scikit-learn, MLflow, and SHAP. This template significantly reduces the time and effort required to build and iterate on ML models, addressing the common challenges of scalability and reproducibility in biomedical research. Adapting our template may save bioinformaticians time in development and enables biomedical researchers to deploy ML projects. Availability and implementation: xML-workFlow is available at https://github.com/MedicalGenomicsLab/xML-workFlow.
翻译:暂无翻译