用少于10行代码训练前沿深度学习新药研发模型

©PaperWeekly · 作者|黄柯鑫
学校|哈佛大学硕士生
研究方向|图学习和生物医疗
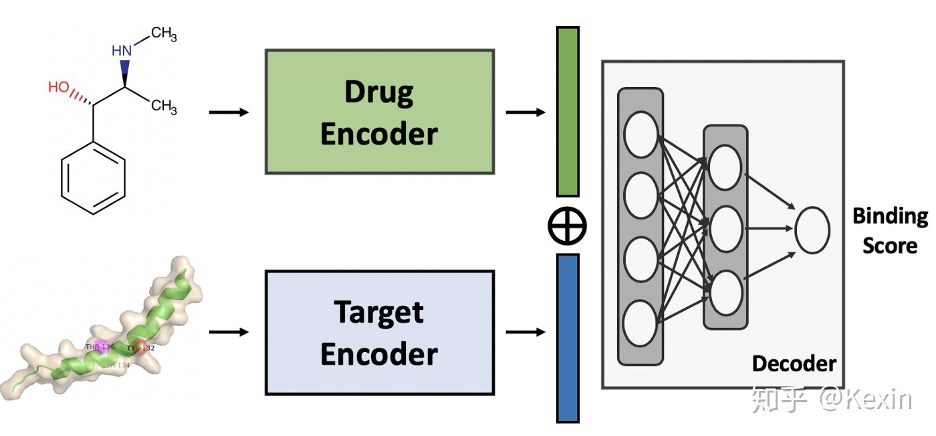
深度学习正在革新药研发行业。在本文中,我们将展示如何使用 DeepPurpose,一个基于 PyTorch 的工具包来解锁 50 多个用于药物-靶标相互作用(Drug-Target Interaction)预测的模型。
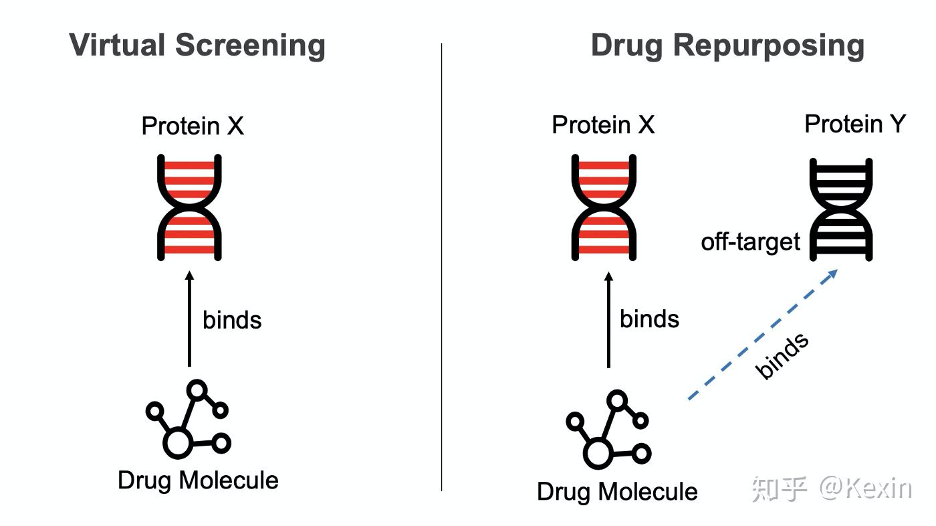
DTI 预测是新药研发中的一项基本任务。DeepPurpose 的操作模式是像 scikit-learn 一样。只需几行代码,就可以利用最前沿的深度学习和药物研发模型。DeepPurpose 还有一个简单的界面来做 DTI 预测的两个重要应用:虚拟筛选(Virtual Screening)和旧药新用(Drug Repurposing)。要了解更多信息,请访问 arxiv 文章和 Github。
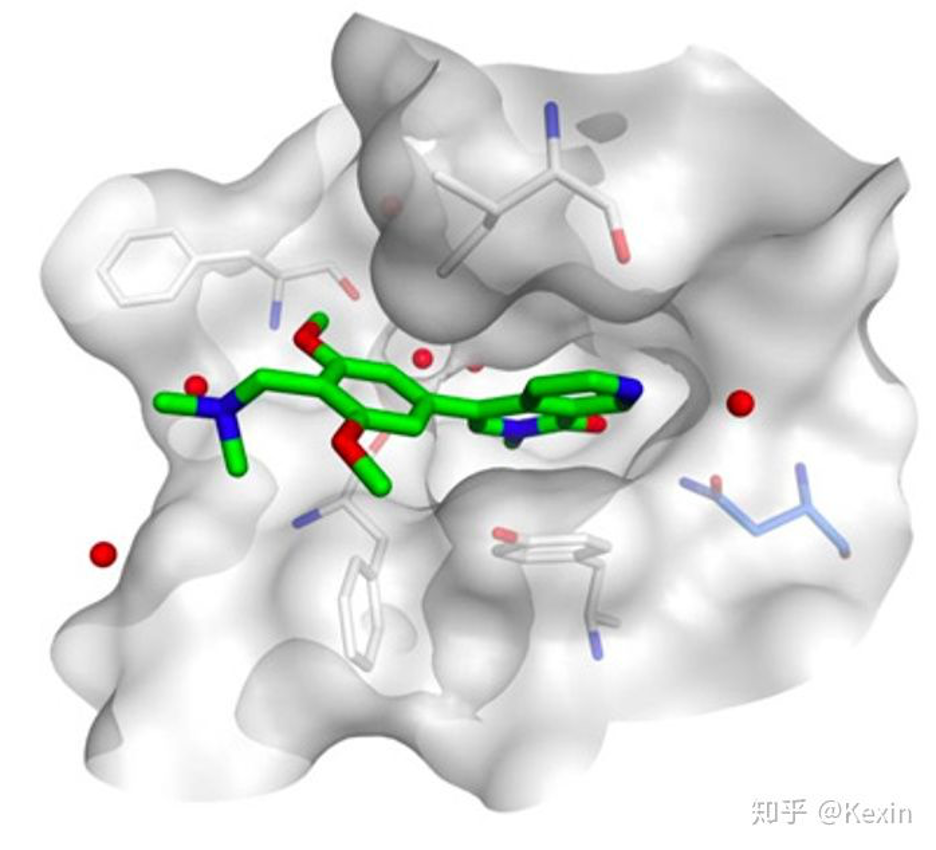
▲ Image by authors.
▲ Image permission granted by Christopher Vakoc.
▲ Image by authors.
▲ Image by authors.
▲ Image by authors.
▲ Image by authors.
▲ Image by authors.
Data loading(数据加载)
Encoder specification(编码器规格)
Data encoding and split(数据编码和分割)
Model configuration generation(模型配置生成)
Model initialization(模型初始化)
Model training(模型训练)
Repurposing/Screening(旧药新用/虚拟筛选)
Model saving and loading(模型保存和加载)
https://github.com/kexinhuang12345/DeepPurpose/blob/master/Tutorial_1_DTI_Prediction.ipynb
https://mybinder.org/v2/gh/kexinhuang12345/DeepPurpose/master
https://github.com/kexinhuang12345/DeepPurpose#install--usage
Data loading(数据加载)
from DeepPurpose import utils, models, dataset
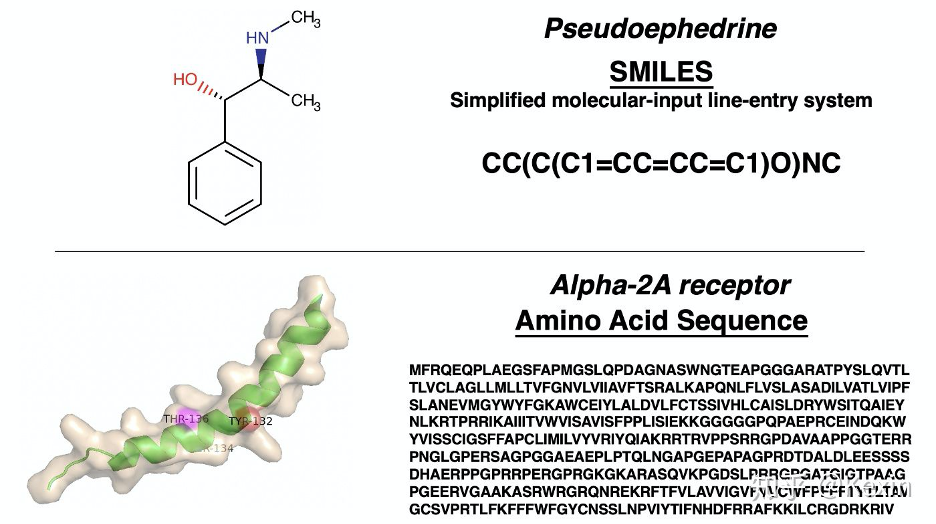
X_drugs, X_targets, y = dataset.load_process_DAVIS(path = './data', binary = False, convert_to_log = True, threshold = 30)
print('Drug 1: ' + X_drugs[0])
print('Target 1: ' + X_targets[0])
print('Score 1: ' + str(y[0]))
# ------ Output ------
# Beginning Processing...
# Beginning to extract zip file...
# Default set to logspace (nM -> p) for easier regression
# Done!
# Drug 1: CC1=C2C=C(C=CC2=NN1)C3=CC(=CN=C3)OCC(CC4=CC=CC=C4)N
# Target 1: MKKFFDSRREQGGSGLGSGSSGGGGSTSGLGSGYIGRVFGIGRQQVTVDEVLAEGGFAIVFLVRTSNGMKCALKRMFVNNEHDLQVCKREIQIMRDLSGHKNIVGYIDSSINNVSSGDVWEVLILMDFCRGGQVVNLMNQRLQTGFTENEVLQIFCDTCEAVARLHQCKTPIIHRDLKVENILLHDRGHYVLCDFGSATNKFQNPQTEGVNAVEDEIKKYTTLSYRAPEMVNLYSGKIITTKADIWALGCLLYKLCYFTLPFGESQVAICDGNFTIPDNSRYSQDMHCLIRYMLEPDPDKRPDIYQVSYFSFKLLKKECPIPNVQNSPIPAKLPEPVKASEAAAKKTQPKARLTDPIPTTETSIAPRQRPKAGQTQPNPGILPIQPALTPRKRATVQPPPQAAGSSNQPGLLASVPQPKPQAPPSQPLPQTQAKQPQAPPTPQQTPSTQAQGLPAQAQATPQHQQQLFLKQQQQQQQPPPAQQQPAGTFYQQQQAQTQQFQAVHPATQKPAIAQFPVVSQGGSQQQLMQNFYQQQQQQQQQQQQQQLATALHQQQLMTQQAALQQKPTMAAGQQPQPQPAAAPQPAPAQEPAIQAPVRQQPKVQTTPPPAVQGQKVGSLTPPSSPKTQRAGHRRILSDVTHSAVFGVPASKSTQLLQAAAAEASLNKSKSATTTPSGSPRTSQQNVYNPSEGSTWNPFDDDNFSKLTAEELLNKDFAKLGEGKHPEKLGGSAESLIPGFQSTQGDAFATTSFSAGTAEKRKGGQTVDSGLPLLSVSDPFIPLQVPDAPEKLIEGLKSPDTSLLLPDLLPMTDPFGSTSDAVIEKADVAVESLIPGLEPPVPQRLPSQTESVTSNRTDSLTGEDSLLDCSLLSNPTTDLLEEFAPTAISAPVHKAAEDSNLISGFDVPEGSDKVAEDEFDPIPVLITKNPQGGHSRNSSGSSESSLPNLARSLLLVDQLIDL
# Score 1: 7.3655227298392685
# --------------------
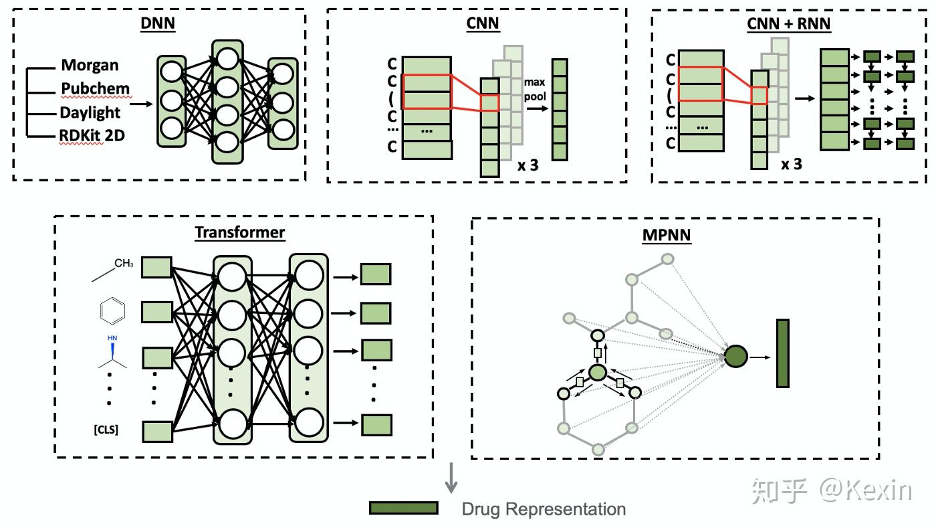
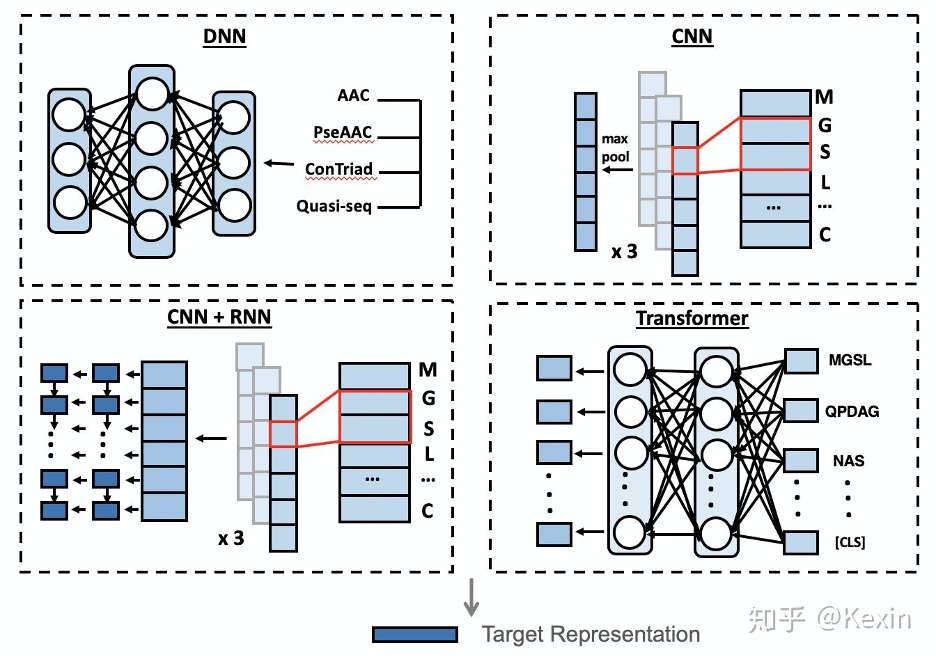
https://github.com/kexinhuang12345/DeepPurpose#encodings
drug_encoding, target_encoding = 'MPNN', 'CNN'
#drug_encoding, target_encoding = 'Morgan', 'Conjoint_triad'Data encoding and split(数据编码和分割)
train, val, test = utils.data_process(X_drugs, X_targets, y,
drug_encoding, target_encoding,
split_method='random',frac=[0.7,0.1,0.2],
random_seed = 1)
# ------ Output ------
# in total: 30056 drug-target pairs
# encoding drug...
# unique drugs: 68
# drug encoding finished...
# encoding protein...
# unique target sequence: 379
# protein encoding finished...
# splitting dataset...
# Done.
# --------------------Model configuration generation(模型配置生成)
https://github.com/kexinhuang12345/DeepPurpose/blob/e169e2f550694145077bb2af95a4031abe400a77/DeepPurpose/utils.py#L486
config = utils.generate_config(drug_encoding = drug_encoding,
target_encoding = target_encoding,
cls_hidden_dims = [1024,1024,512],
train_epoch = 5,
LR = 0.001,
batch_size = 128,
hidden_dim_drug = 128,
mpnn_hidden_size = 128,
mpnn_depth = 3,
cnn_target_filters = [32,64,96],
cnn_target_kernels = [4,8,12]
)
Model initialization(模型初始化)
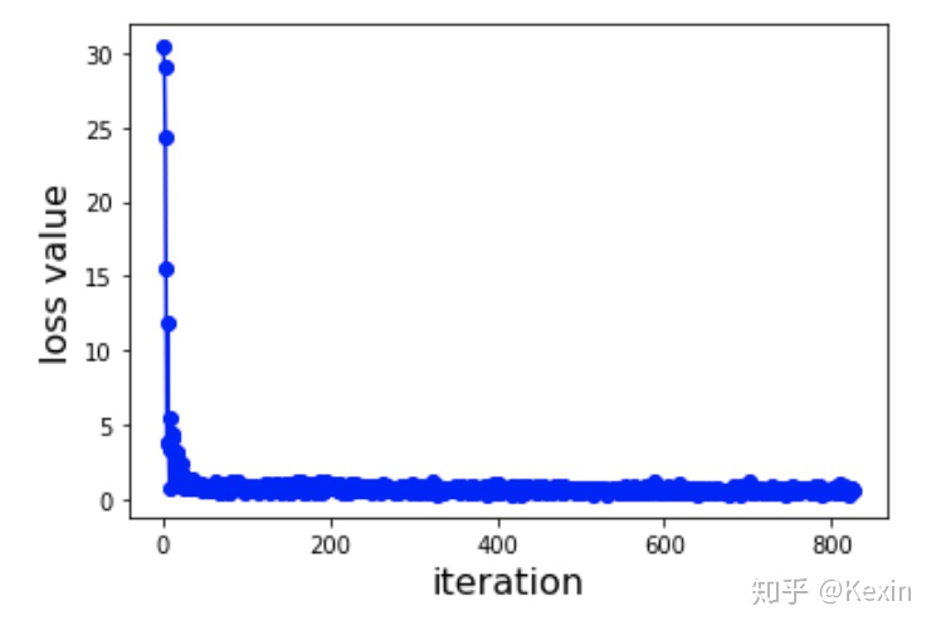
model = models.model_initialize(**config)
model.train(train, val, test)
# ------ Output ------
# Let's use 1 GPU!
# --- Data Preparation ---
# --- Go for Training ---
#Training at Epoch 1 iteration 0 with loss 30.3964. Total time 0.0 hours
# Training at Epoch 1 iteration 100 with loss 0.59583. Total time 0.01277 hours
# Validation at Epoch 1 , MSE: 0.79965 , Pearson Correlation: 0.34392 with p-value: 3.23317 , Concordance Index: 0.67805
# .....
# --- Go for Testing ---
# Testing MSE: 0.5567689292029449 , Pearson Correlation: 0.5585790614300189 with p-value: 0.0 , Concordance Index: 0.783525396597284
# --- Training Finished ---
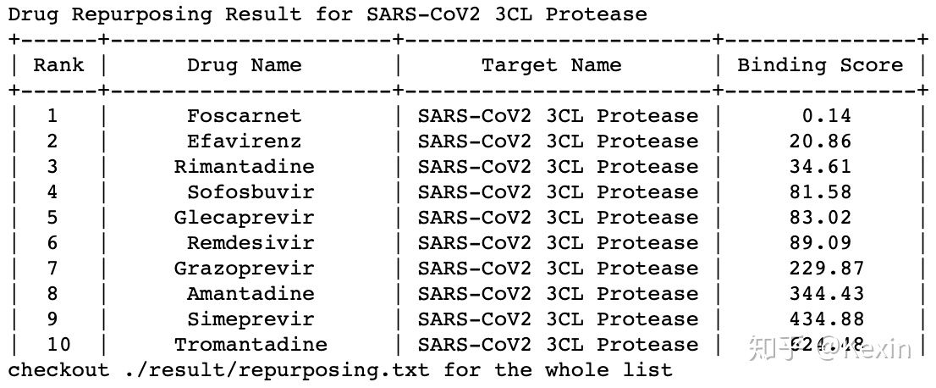
# --------------------target, target_name = dataset.load_SARS_CoV2_Protease_3CL()
repurpose_drugs, repurpose_drugs_name, repurpose_drugs_pubchem_cid = dataset.load_antiviral_drugs()
y_pred = models.repurpose(X_repurpose = repurpose_drugs, target = target, model = model,
drug_names = repurpose_drugs_name, target_name = target_name,
result_folder = "./result/", convert_y = True)
# ------ Output ------
# repurposing...
# in total: 82 drug-target pairs
# encoding drug...
# unique drugs: 81
# drug encoding finished...
# encoding protein...
# unique target sequence: 1
# protein encoding finished...
# Done.
# predicting...
# --------------------
target, drugs = dataset.load_IC50_1000_Samples()
y_pred = models.virtual_screening(drugs, target, model)
# ------ Output ------
# virtual screening...
# in total: 100 drug-target pairs
# encoding drug...
# unique drugs: 100
# drug encoding finished...
# encoding protein...
# unique target sequence: 93
# protein encoding finished...
# Done.
# predicting...
# --------------------
model.save_model('./tutorial_model')
model = models.model_pretrained(path_dir = './tutorial_model')https://github.com/kexinhuang12345/DeepPurpose#pretrained-models
model = models.model_pretrained(model = 'MPNN_CNN_BindingDB')
# ------ Output ------
# Beginning Downloading MPNN_CNN_BindingDB Model...
# Downloading finished... Beginning to extract zip file...
# pretrained model Successfully Downloaded...
# --------------------https://github.com/kexinhuang12345/DeepPurpose/blob/master/DEMO/Drug_Property_Pred-Ax-Hyperparam-Tune.ipynb
from DeepPurpose import utils, models, dataset, property_pred
X_drugs, X_targets, y = dataset.load_AID1706_SARS_CoV_3CL(path = './data', binary = True, threshold = 15, balanced = True)
drug_encoding = 'MPNN'
train, val, test = utils.data_process(X_drug = X_drugs, y = y, drug_encoding = drug_encoding,
split_method='random',frac=[0.7,0.1,0.2],
random_seed = 1)
config = utils.generate_config(drug_encoding = drug_encoding,
cls_hidden_dims = [1024,1024,512],
train_epoch = 5,
LR = 0.001,
batch_size = 128,
hidden_dim_drug = 128,
mpnn_hidden_size = 128,
mpnn_depth = 3
)
model = property_pred.model_initialize(**config)
model.train(train, val, test)
https://github.com/kexinhuang12345/DeepPurpose/blob/master/Tutorial_1_DTI_Prediction.ipynb
https://github.com/kexinhuang12345/DeepPurpose/blob/master/Tutorial_2_Drug_Property_Pred_Assay_Data.ipynb
总结
参考文献
[1] Mullard, A. New drugs cost US$2.6 billion to develop. Nature Reviews Drug Discovery (2014).
[2] Fleming, Nic. How artificial intelligence is changing drug discovery. Nature (2018).
[3] Smalley, E. AI-powered drug discovery captures pharma interest. Nature Biotechnology (2017).
[4] Gschwend DA, Good AC, Kuntz ID. Molecular docking towards drug discovery. Journal of Molecular Recognition: An Interdisciplinary Journal (1996).
[5] Mayr, Andreas, et al. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chemical science (2018).
[6] Öztürk H, Özgür A, Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics (2018).
[7] Nguyen, Thin, Hang Le, and Svetha Venkatesh. GraphDTA: prediction of drug–target binding affinity using graph convolutional networks. BioRxiv (2019).
[8] Tsubaki, Masashi, Kentaro Tomii, and Jun Sese. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics (2019).
[9] Lee, Ingoo, Jongsoo Keum, and Hojung Nam. DeepConv-DTI: Prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS computational biology (2019).
[10] Chen, Xing, et al. Drug–target interaction prediction: databases, web servers and computational models. Briefings in bioinformatics (2016).
[11] Huang K, Xiao C, Hoang T, Glass L, Sun J. CASTER: Predicting Drug Interactions with Chemical Substructure Representation. AAAI (2020).
更多阅读



#投 稿 通 道#
让你的论文被更多人看到
如何才能让更多的优质内容以更短路径到达读者群体,缩短读者寻找优质内容的成本呢?答案就是:你不认识的人。
总有一些你不认识的人,知道你想知道的东西。PaperWeekly 或许可以成为一座桥梁,促使不同背景、不同方向的学者和学术灵感相互碰撞,迸发出更多的可能性。
PaperWeekly 鼓励高校实验室或个人,在我们的平台上分享各类优质内容,可以是最新论文解读,也可以是学习心得或技术干货。我们的目的只有一个,让知识真正流动起来。
📝 来稿标准:
• 稿件确系个人原创作品,来稿需注明作者个人信息(姓名+学校/工作单位+学历/职位+研究方向)
• 如果文章并非首发,请在投稿时提醒并附上所有已发布链接
• PaperWeekly 默认每篇文章都是首发,均会添加“原创”标志
📬 投稿邮箱:
• 投稿邮箱:hr@paperweekly.site
• 所有文章配图,请单独在附件中发送
• 请留下即时联系方式(微信或手机),以便我们在编辑发布时和作者沟通
🔍
现在,在「知乎」也能找到我们了
进入知乎首页搜索「PaperWeekly」
点击「关注」订阅我们的专栏吧
关于PaperWeekly
PaperWeekly 是一个推荐、解读、讨论、报道人工智能前沿论文成果的学术平台。如果你研究或从事 AI 领域,欢迎在公众号后台点击「交流群」,小助手将把你带入 PaperWeekly 的交流群里。