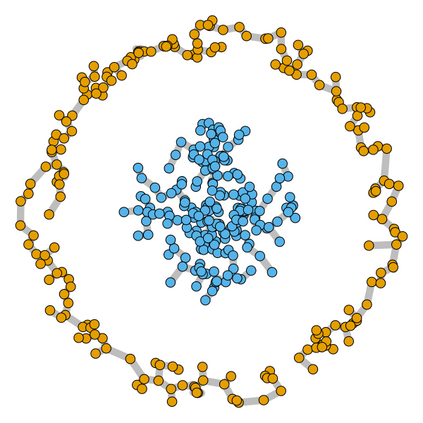
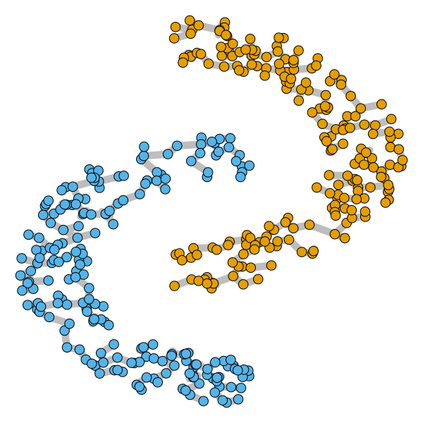
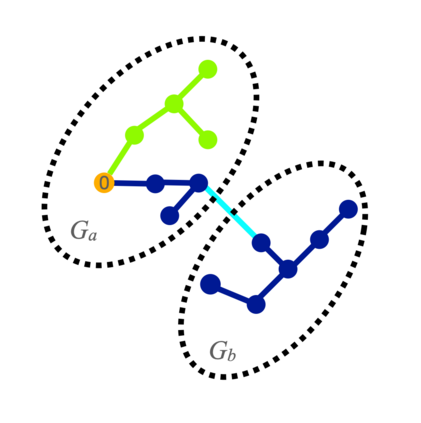
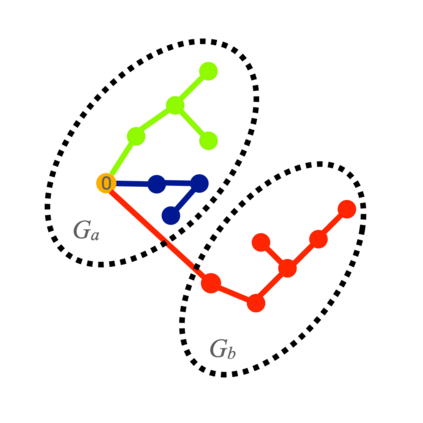
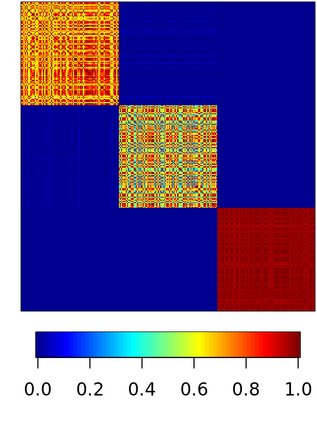
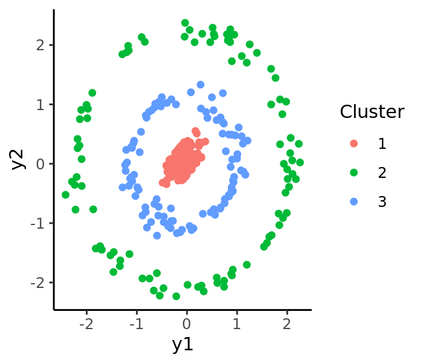
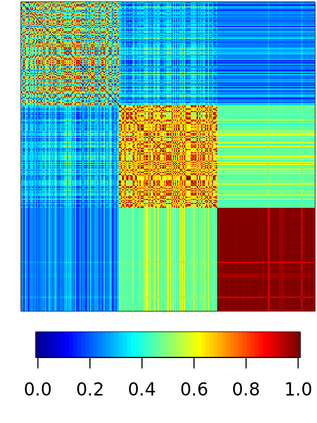
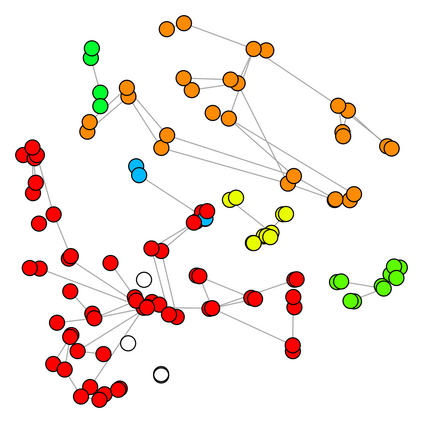
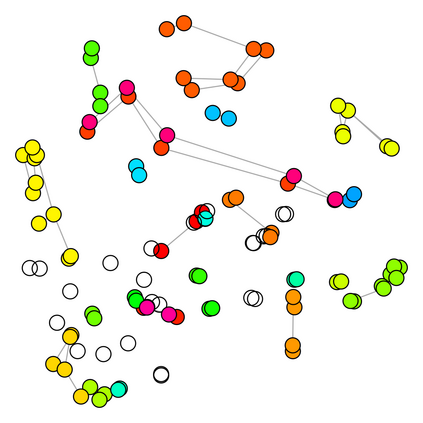
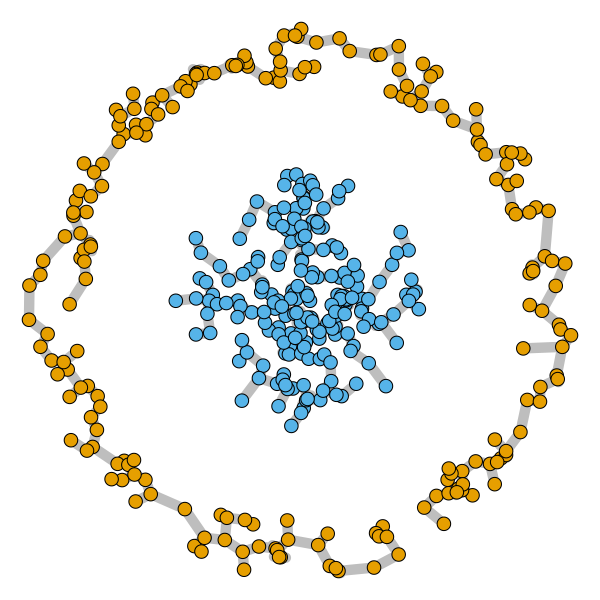
Spectral clustering algorithms are very popular. Starting from a pairwise similarity matrix, spectral clustering gives a partition of data that approximately minimizes the total similarity scores across clusters. Since there is no need to model how data are distributed within each cluster, such a method enjoys algorithmic simplicity and robustness in clustering non-Gaussian data such as those near manifolds. Nevertheless, several important questions are unaddressed, such as how to estimate the similarity scores and cluster assignment probabilities, as important uncertainty estimates in clustering. In this article, we propose to solve these problems with a discovered generative modeling counterpart. Our clustering model is based on a spanning forest graph that consists of several disjoint spanning trees, with each tree corresponding to a cluster. Taking a Bayesian approach, we assign proper densities on the root and leaf nodes, and we prove that the posterior mode is almost the same as spectral clustering estimates. Further, we show that the associated generative process, named "forest process", is a continuous extension to the classic urn process, hence inheriting many nice properties such as having unbounded support for the number of clusters and being amenable to existing partition probability function; at the same time, we carefully characterize their differences. We demonstrate a novel application in joint clustering of multiple-subject functional magnetic resonance imaging scans of the human brain.
翻译:光谱群集算法非常流行。 从相近的相近群集矩阵开始, 光谱群集可以分出一个可以将各组群之间完全相似的分数缩小到最小的数据。 由于不需要在每组群中模拟数据是如何分配的, 这种方法在将非古裔数据( 如临近的多元体) 组合起来时, 具有算法上的简单性和稳健性。 然而, 有几个重要问题没有解决, 比如如何估算相似分数和群集分配概率, 作为群集中重要的不确定性估计。 在文章中, 我们提议用一个发现的基因模型来解决这些问题。 我们的群集模型模型基于一个覆盖森林的图, 由几棵不相连的树组成, 每棵树对应一个组群。 采取巴伊西亚方法, 我们给根和叶节点等非古裔数据分配适当的密度, 我们证明外表模式与光谱群集估计值几乎相同。 此外, 我们证明相关的基因组化过程, 叫做“ 森林过程” 是典型的延续过程, 从而继承了许多好的属性图象学属性, 例如, 具有一些不相连的功能类组群集的模型 。 我们在 的模型中 的模型化中, 正在展示着 的 的模型的模型的模型的模型的模型的模型的模型 。