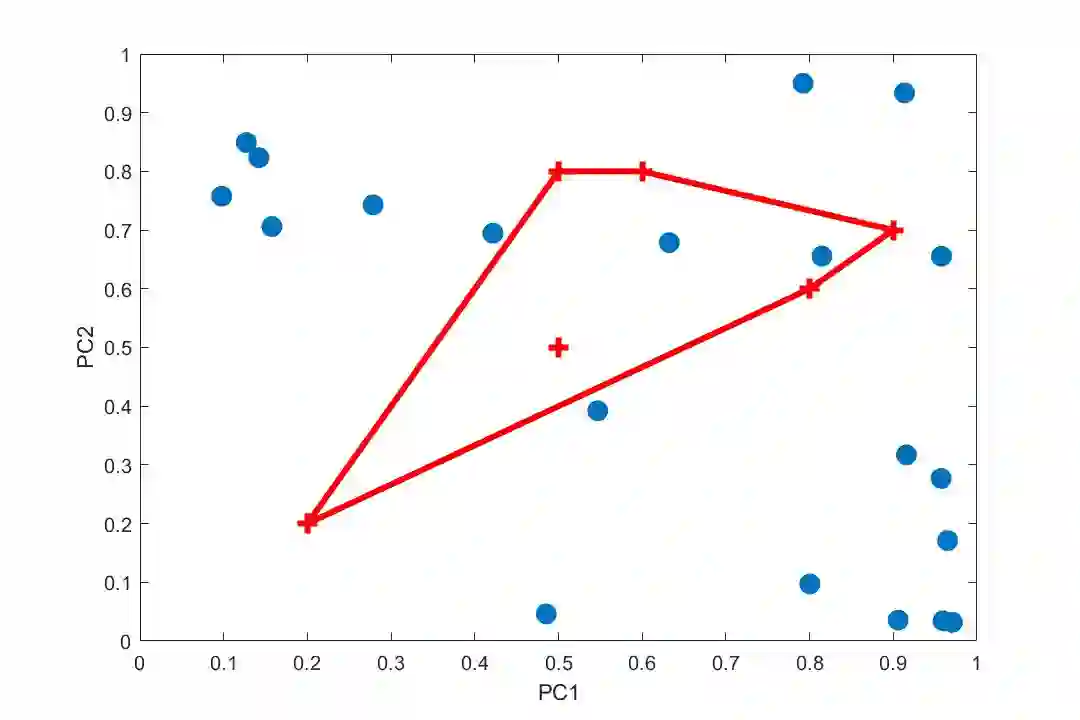
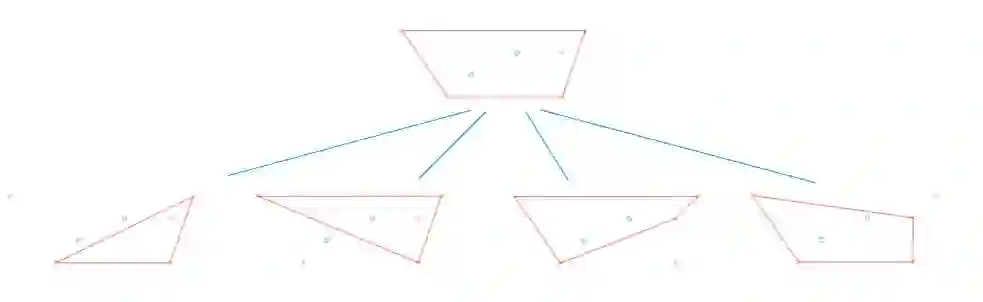
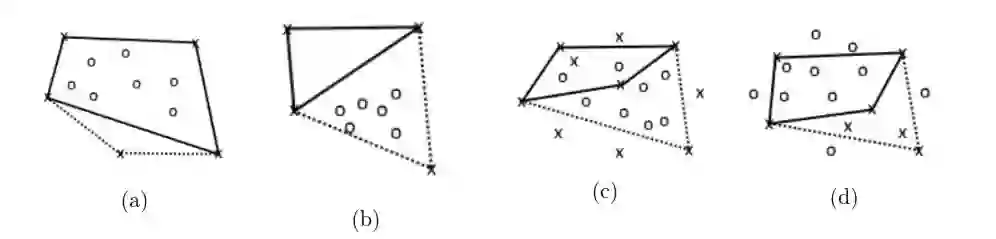
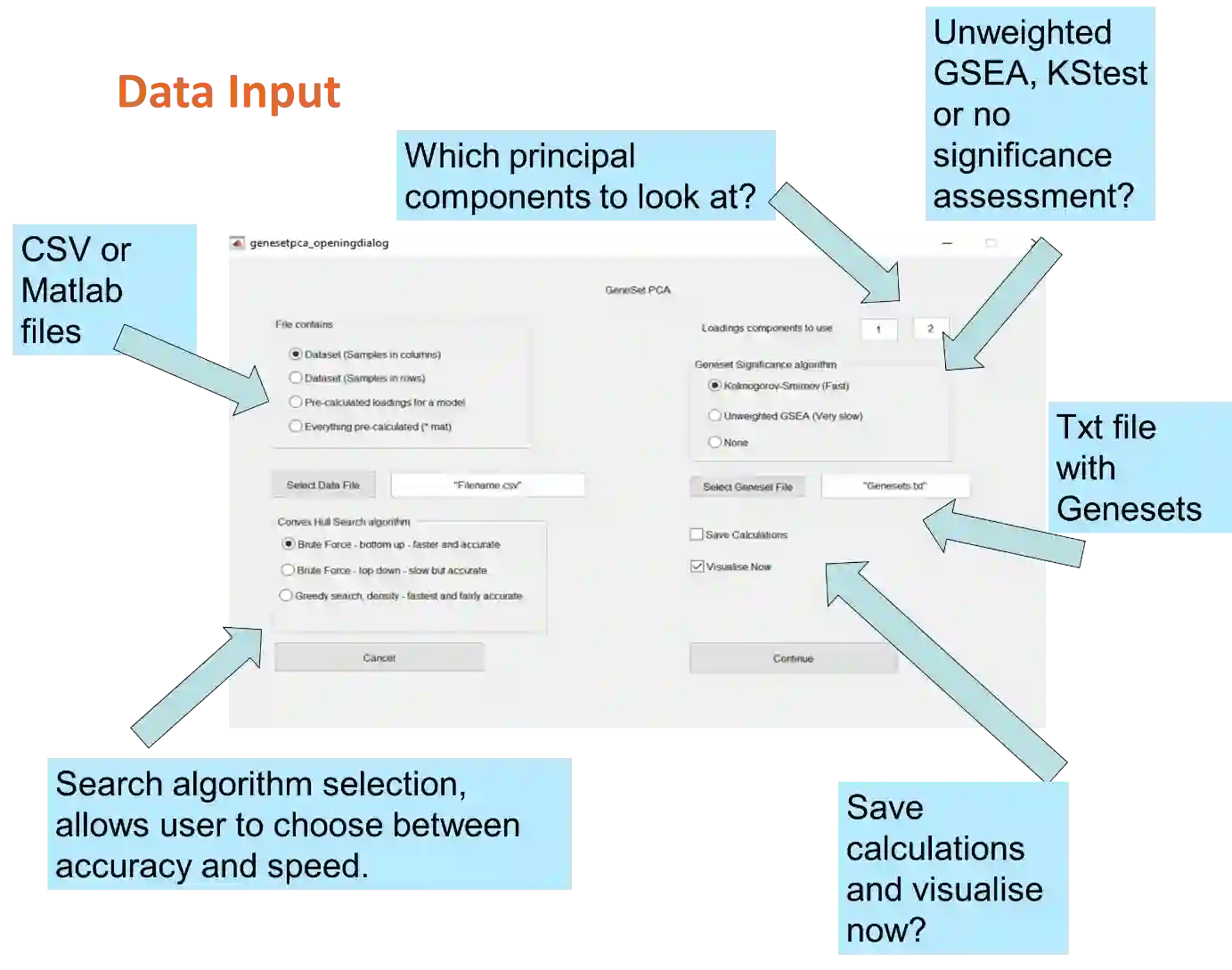
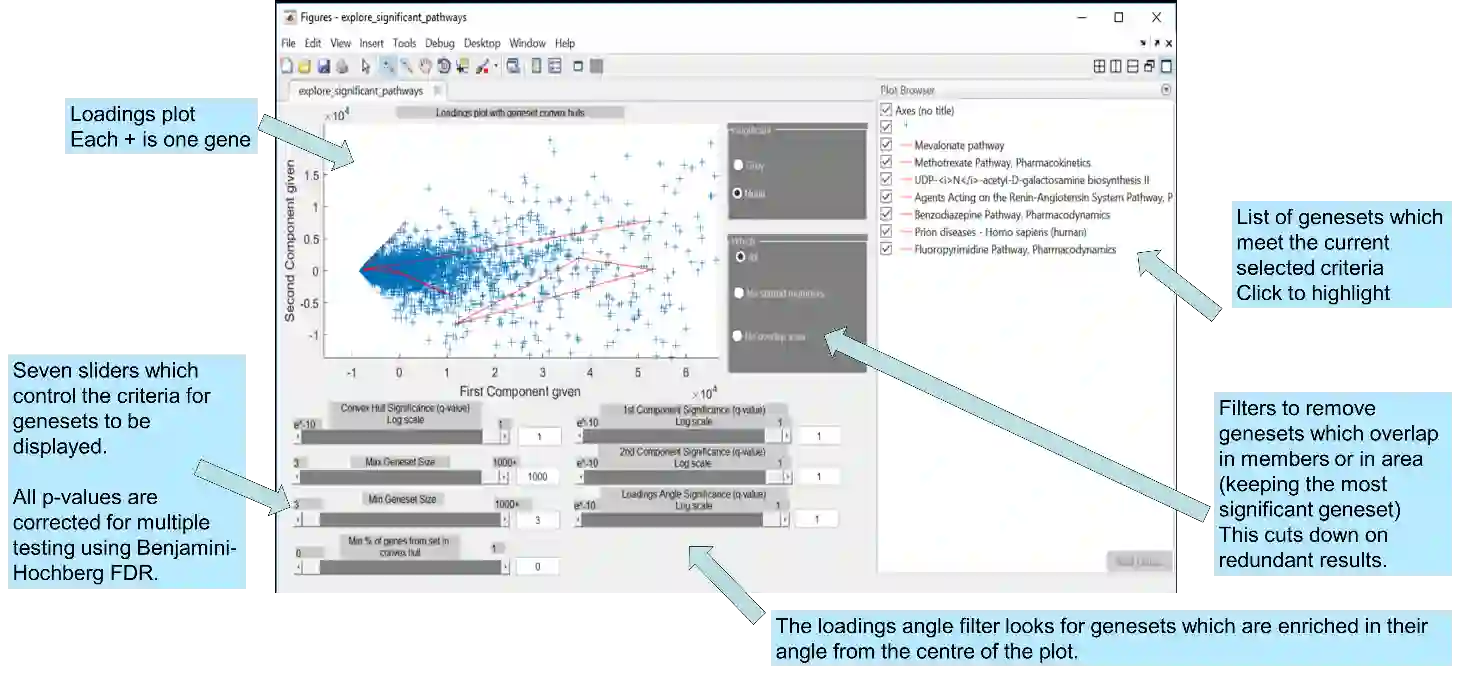
Principal Component Analysis (PCA) and other multi-variate models are often used in the analysis of "omics" data. These models contain much information which is currently neither easily accessible nor interpretable. Here we present an algorithmic method which has been developed to integrate this information with existing databases of background knowledge, stored in the form of known sets (for instance genesets or pathways). To make this accessible we have produced a Graphical User Interface (GUI) in Matlab which allows the overlay of known set information onto the loadings plot and thus improves the interpretability of the multi-variate model. For each known set the optimal convex hull, covering a subset of elements from the known set, is found through a search algorithm and displayed. In this paper we discuss two main topics; the details of the search algorithm for the optimal convex hull for this problem and the GUI interface which is freely available for download for academic use.
翻译:分析“ 组合” 数据时经常使用主元件分析( PCA) 和其他多变量模型。 这些模型包含许多目前不易获取和解释的信息。 我们在这里展示了一种算法方法, 将这些信息与现有的背景知识数据库整合起来, 以已知的数据集( 例如基因集或路径) 的形式存储。 为了便于使用, 我们在 Matlab 制作了一个图形用户界面( GUI ), 允许将已知的数据集信息覆盖到加载图中, 从而改进多变量模型的可解释性。 对于每个已知的模型来说, 包含已知集的一组元素的最佳的螺旋体, 是通过搜索算法找到并展示的。 在本文中, 我们讨论两个主要主题; 这一问题的最佳矩形体搜索算法的细节, 以及可自由下载供学术使用的图形界面界面 。