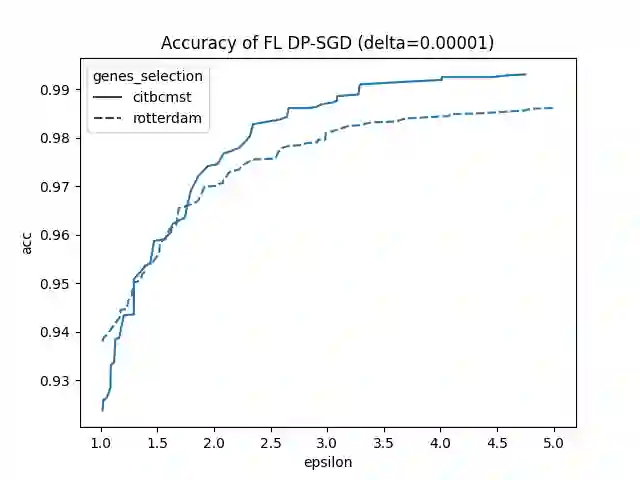
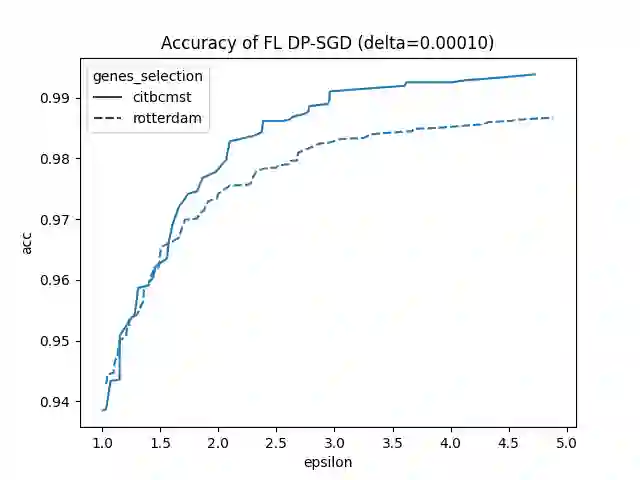
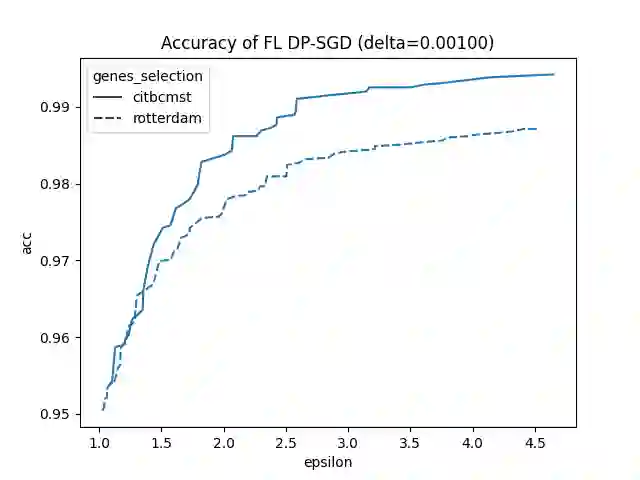
Since 2014, the NIH funded iDASH (integrating Data for Analysis, Anonymization, SHaring) National Center for Biomedical Computing has hosted yearly competitions on the topic of private computing for genomic data. For one track of the 2020 iteration of this competition, participants were challenged to produce an approach to federated learning (FL) training of genomic cancer prediction models using differential privacy (DP), with submissions ranked according to held-out test accuracy for a given set of DP budgets. More precisely, in this track, we are tasked with training a supervised model for the prediction of breast cancer occurrence from genomic data split between two virtual centers while ensuring data privacy with respect to model transfer via DP. In this article, we present our 3rd place submission to this competition. During the competition, we encountered two main challenges discussed in this article: i) ensuring correctness of the privacy budget evaluation and ii) achieving an acceptable trade-off between prediction performance and privacy budget.
翻译:自2014年以来,NIH资助了国家生物医学计算中心(IDASH) iDASH(综合分析、匿名、SHaring数据综合数据),每年举办关于基因组数据私人计算主题的年度竞赛,对于2020年这一竞争迭代的一个方面,参与者受到挑战,要制定一种方法,利用不同的隐私(DP)对基因组癌预测模型进行联合学习(FL)培训,提交材料按特定一组DP预算的稳妥测试准确性排列。更确切地说,在这一方面,我们的任务是培训一个监督模型,用于预测两个虚拟中心通过基因组数据分割的乳腺癌发生率,同时确保通过DP进行模型转移时的数据隐私。在本条中,我们介绍了我们向这一竞争提交的第3位材料。在竞争期间,我们遇到了该条讨论的两个主要挑战:一) 确保隐私预算评估的正确性,二) 在预测性业绩和隐私预算之间实现可接受的权衡。