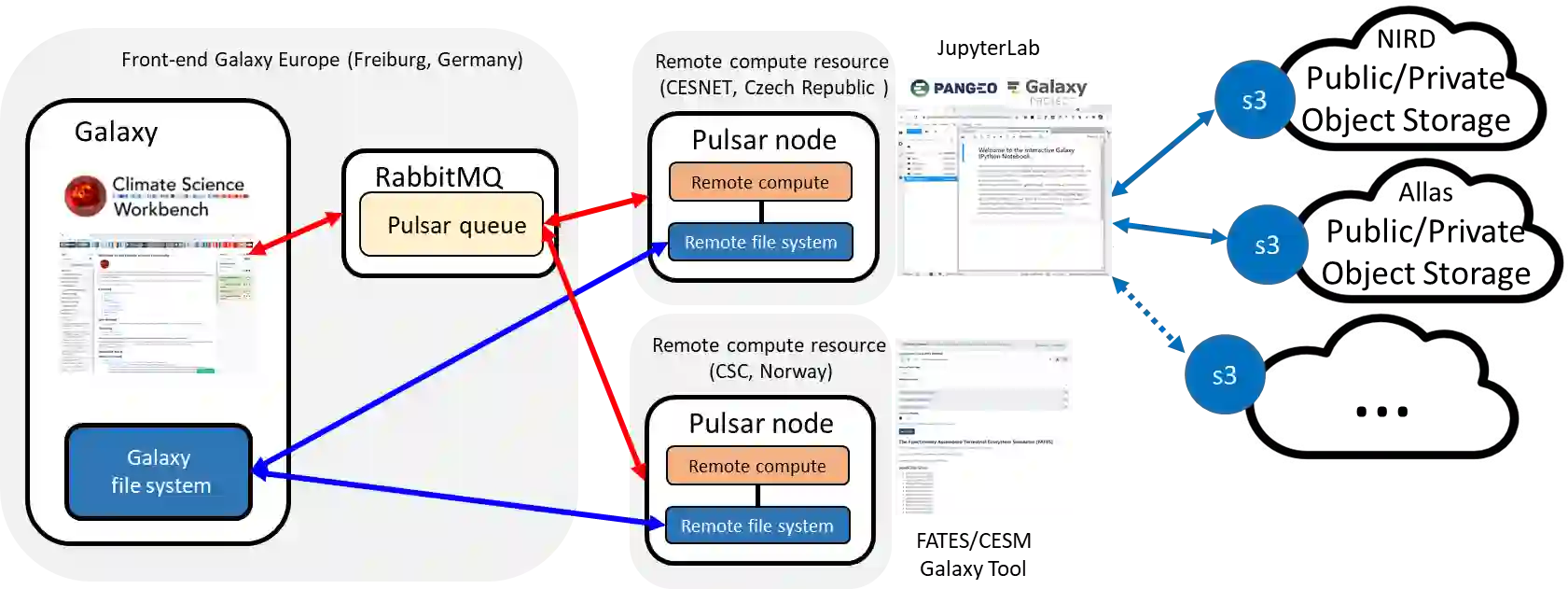
To reproduce eScience, several challenges need to be solved: scientific workflows need to be automated; the involved software versions need to be provided in an unambiguous way; input data needs to be easily accessible; High-Performance Computing (HPC) clusters are often involved and to achieve bit-to-bit reproducibility, it might be even necessary to execute the code on a particular cluster to avoid differences caused by different HPC platforms (and unless this is a scientist's local cluster, it needs to be accessed across (administrative) borders). Preferably, to allow even inexperienced users to (re-)produce results, all should be user-friendly. While some easy-to-use web-based scientific portals support already to access HPC resources, this typically only refers to computing and data resources that are local. By the example of two community-specific portals in the fields of biodiversity and climate research, we present a solution for accessing remote HPC (and cloud) compute and data resources from scientific portals across borders, involving rigorous container-based packaging of the software version and setup automation, thus enhancing reproducibility.
翻译:为了复制电子科学,需要解决若干挑战:科学工作流程需要自动化;需要以明确的方式提供所涉软件版本;投入数据需要容易获取; 高性能计算(HPC)群集经常参与进来,并实现位对位复制,因此,也许甚至有必要对特定组群执行守则,以避免不同的HPC平台造成的差异(除非这是一个科学家的本地群集,它需要跨(行政)边界进入)。为了让甚至没有经验的用户能够(重新)产生结果,所有软件都应方便用户使用。虽然一些基于网络的容易使用的网络科学门户已经支持获取HPC资源,但通常仅指计算机和当地的数据资源。我们以生物多样性和气候研究领域的两个社区专用门户为例,提出了从跨界科学门户获取远程HPC(和云)计算和数据资源的解决办法,包括软件版本的严格集装箱包装和设置自动化,从而加强了再分析能力。