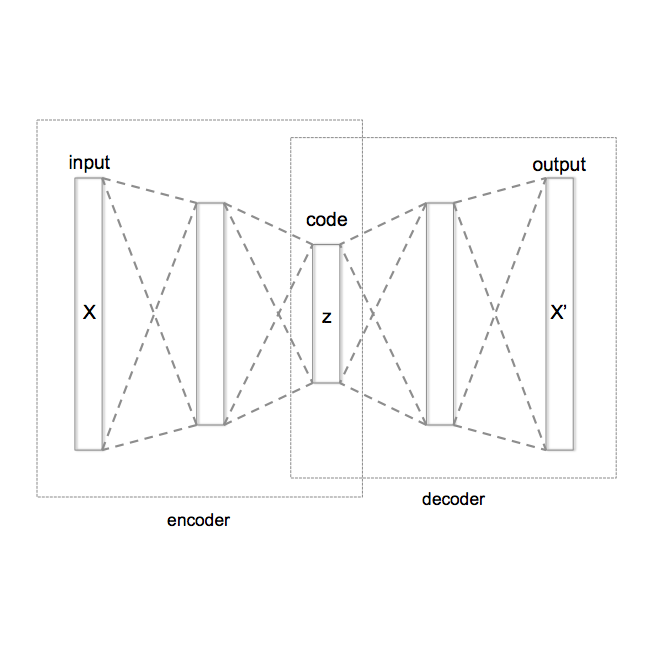
Barrett's Esophagus (BE) is the only precursor known to Esophageal Adenocarcinoma (EAC), a type of esophageal cancer with poor prognosis upon diagnosis. Therefore, diagnosing BE is crucial in preventing and treating esophageal cancer. While supervised machine learning supports BE diagnosis, high interobserver variability in histopathological training data limits these methods. Unsupervised representation learning via Variational Autoencoders (VAEs) shows promise, as they map input data to a lower-dimensional manifold with only useful features, characterizing BE progression for improved downstream tasks and insights. However, the VAE's Euclidean latent space distorts point relationships, hindering disease progression modeling. Geometric VAEs provide additional geometric structure to the latent space, with RHVAE assuming a Riemannian manifold and $\mathcal{S}$-VAE a hyperspherical manifold. Our study shows that $\mathcal{S}$-VAE outperforms vanilla VAE with better reconstruction losses, representation classification accuracies, and higher-quality generated images and interpolations in lower-dimensional settings. By disentangling rotation information from the latent space, we improve results further using a group-based architecture. Additionally, we take initial steps towards $\mathcal{S}$-AE, a novel autoencoder model generating qualitative images without a variational framework, but retaining benefits of autoencoders such as stability and reconstruction quality.
翻译:暂无翻译