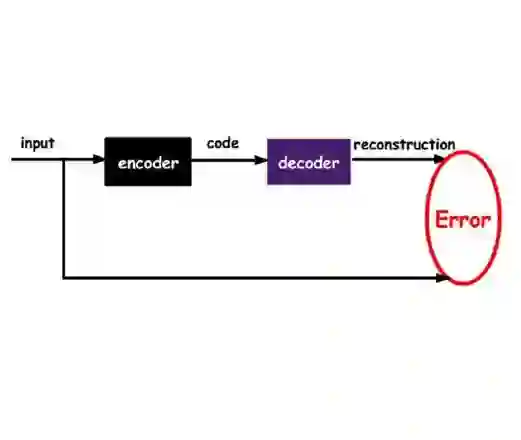
3D ultrasound (3DUS) becomes more interesting for target tracking in radiation therapy due to its capability to provide volumetric images in real-time without using ionizing radiation. It is potentially usable for tracking without using fiducials. For this, a method for learning meaningful representations would be useful to recognize anatomical structures in different time frames in representation space (r-space). In this study, 3DUS patches are reduced into a 128-dimensional r-space using conventional autoencoder, variational autoencoder and sliced-wasserstein autoencoder. In the r-space, the capability of separating different ultrasound patches as well as recognizing similar patches is investigated and compared based on a dataset of liver images. Two metrics to evaluate the tracking capability in the r-space are proposed. It is shown that ultrasound patches with different anatomical structures can be distinguished and sets of similar patches can be clustered in r-space. The results indicate that the investigated autoencoders have different levels of usability for target tracking in 3DUS.
翻译:3D超声波(3DUS)对于辐射疗法的目标跟踪更加有趣,因为它有能力不使用电离辐射而实时提供体积图象。它可能用于不使用电离辐射进行跟踪。为此,学习有意义的表达方法将有益于识别代表空间(r-space)不同时间框架内的解剖结构。在本研究中,3DUS补丁将使用常规自动coder、变异自动coder和切片瓦瑟斯坦自动编码器,缩小为128维R-空间。在R空间中,根据肝脏图象数据集,对分离不同超声波补丁和识别类似补丁的能力进行调查和比较。提出了评价R-空间跟踪能力的两种衡量标准,显示不同解剖结构的超声波补丁可以区分,在R-空间中也可以将相似的补丁组合组合。结果显示,所调查的自动分解器在3DUS中具有不同水平的目标跟踪可用性。