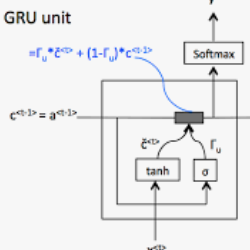
The work for predicting drug and target affinity(DTA) is crucial for drug development and repurposing. In this work, we propose a novel method called GDGRU-DTA to predict the binding affinity between drugs and targets, which is based on GraphDTA, but we consider that protein sequences are long sequences, so simple CNN cannot capture the context dependencies in protein sequences well. Therefore, we improve it by interpreting the protein sequences as time series and extracting their features using Gate Recurrent Unit(GRU) and Bidirectional Gate Recurrent Unit(BiGRU). For the drug, our processing method is similar to that of GraphDTA, but uses two different graph convolution methods. Subsequently, the representation of drugs and proteins are concatenated for final prediction. We evaluate the proposed model on two benchmark datasets. Our model outperforms some state-of-the-art deep learning methods, and the results demonstrate the feasibility and excellent feature capture ability of our model.
翻译:预测药物和目标亲近性(DTA)的工作对于药物发展和重新定位至关重要。在这项工作中,我们提出一种名为GDGRU-DTA的新方法,以预测药物和目标之间的约束性亲近性,该方法以GIDDA为基础,但我们认为蛋白质序列是长序列,因此简单的CNN无法很好地捕捉蛋白序列的上下文依赖性。因此,我们通过将蛋白序列解读为时间序列,并利用Gate经常项目(GRU)和双向门经常项目(BIGRU)提取其特征来改进蛋白序列。关于药物,我们的加工方法与GIDTA相似,但使用两种不同的图解方法。随后,药物和蛋白的表示方式被组合为最终预测。我们评价了两个基准数据集的拟议模型。我们的模型超越了一些最先进的深层次学习方法,结果显示了我们模型的可行性和极好的特征采集能力。