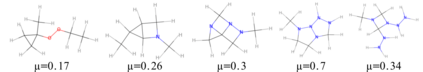
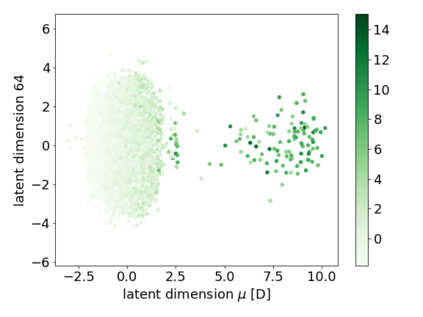
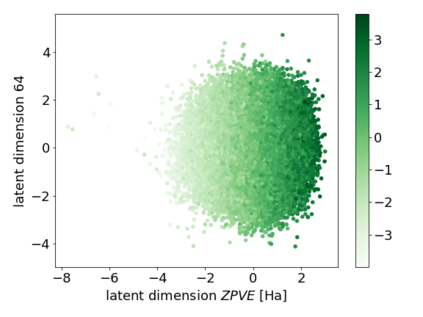
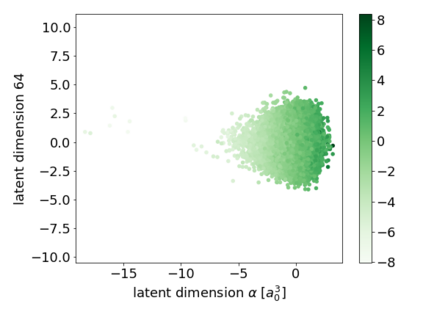
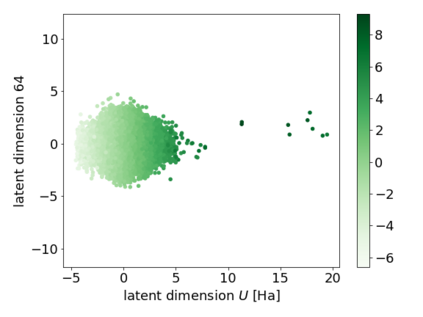
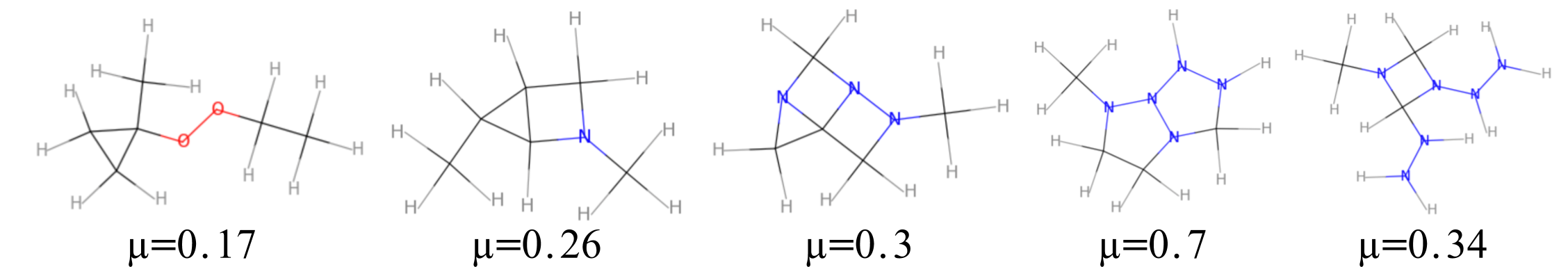
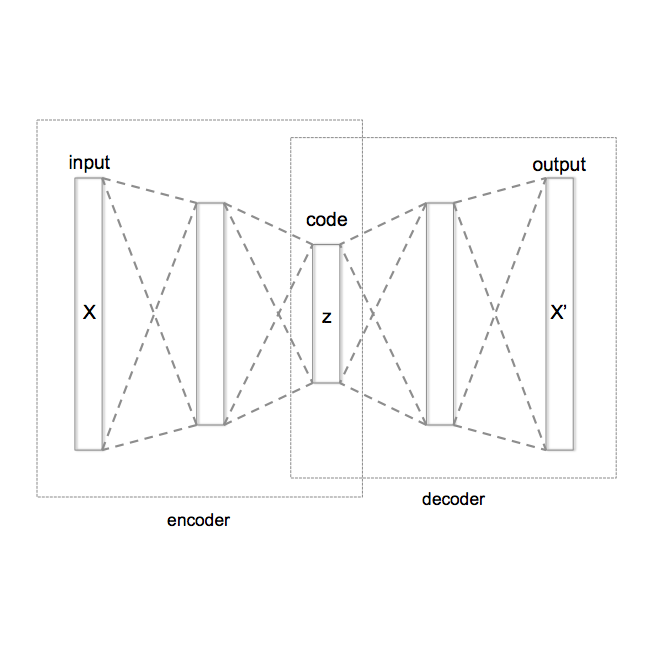
We propose a combination of a variational autoencoder and a transformer based model which fully utilises graph convolutional and graph pooling layers to operate directly on graphs. The transformer model implements a novel node encoding layer, replacing the position encoding typically used in transformers, to create a transformer with no position information that operates on graphs, encoding adjacent node properties into the edge generation process. The proposed model builds on graph generative work operating on graphs with edge features, creating a model that offers improved scalability with the number of nodes in a graph. In addition, our model is capable of learning a disentangled, interpretable latent space that represents graph properties through a mapping between latent variables and graph properties. In experiments we chose a benchmark task of molecular generation, given the importance of both generated node and edge features. Using the QM9 dataset we demonstrate that our model performs strongly across the task of generating valid, unique and novel molecules. Finally, we demonstrate that the model is interpretable by generating molecules controlled by molecular properties, and we then analyse and visualise the learned latent representation.
翻译:我们提议将一个变异自动编码器和一个基于变压器的模型结合起来,该模型充分利用图层的变相和图形集合层,直接在图形上运行。变压器模型采用一个新的节点编码层,取代变压器通常使用的定位编码,以创建一个没有在图形上运行的定位信息的变压器,将相邻节点特性编码到边缘生成过程中。拟议模型以具有边缘特征的图形操作的图解基因化工作为基础,创造一个模型,与图表中的节点数相比,能够提供更好的可调整性。此外,我们的模型能够通过潜在变量和图形属性之间的映射来学习一个分解、可解释的显示图形属性的潜在空间。在实验中,我们选择了分子生成的基准任务,因为两者都产生了节点和边缘特性。使用 QM9 数据集,我们证明我们的模型在产生有效、 独特和新颖的分子的任务中表现得力很强。最后,我们证明模型可以通过生成分子特性控制的分子来解释,然后我们分析并直视所学的潜表。