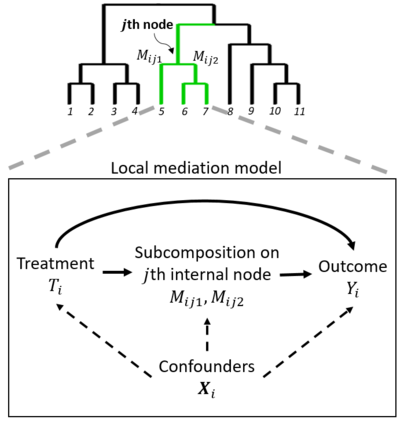
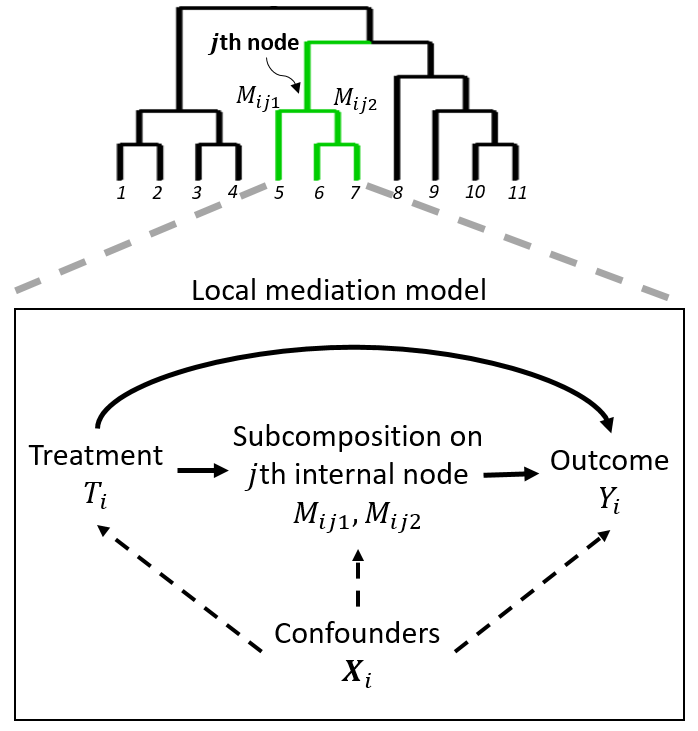
Recent studies suggest that the microbiome can be an important mediator in the effect of a treatment on an outcome. Microbiome data generated from sequencing experiments contain the relative abundance of a large number of microbial taxa with their evolutionary relationships represented by a phylogenetic tree. The compositional and high-dimensional nature of the microbiome mediator invalidates standard mediation analyses. We propose a phylogeny-based mediation analysis method (PhyloMed) for the microbiome mediator. PhyloMed models the microbiome mediation effect through a cascade of independent local mediation models on the internal nodes of the phylogenetic tree. Each local model captures the mediation effect of a subcomposition at a given taxonomic resolution. The method improves the power of the mediation test by enriching weak and sparse signals across mediating taxa that tend to cluster on the tree. In each local model, we further empower PhyloMed by using a mixture distribution to obtain the subcomposition mediation test p-value, which takes into account the composite nature of the null hypothesis. PhyloMed enables us to test the overall mediation effect of the entire microbial community and pinpoint internal nodes with significant subcomposition mediation effects. Our extensive simulations demonstrate the validity of PhyloMed and its substantial power gain over existing methods. An application to a real study further showcases the advantages of our method.
翻译:最近的研究显示,微生物可能是对结果进行治疗的一个重要调解者。从测序实验中产生的微生物数据包含大量微生物分类及其由植物基因树代表的进化关系的相对丰度。微生物调解者的组成和高度性质使得标准的调解分析无效。我们建议微生物调解者采用基于植物的调解分析方法(PhyloMed),通过一系列独立的当地调解模型对植物基因树的内部节点产生分化效应。每个地方模型都捕捉了在特定分类解析分辨率上进行子组合的调解效应。该方法通过在往往集中在树上的媒介分类中丰富薄弱和稀少的信号,提高了调解测试的力量。我们在每个地方模型中,我们进一步通过混合分布来增强PhyloMed的调解方法(PhyloMed),以获得子组合调解测试 pvalueferation pvality,这考虑到了无效假设的复合性质。PhyloMed使我们得以测试在特定分类解析分辨率解析方法上的总体调解效果,并展示了整个微缩方法的优势。
相关内容
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem