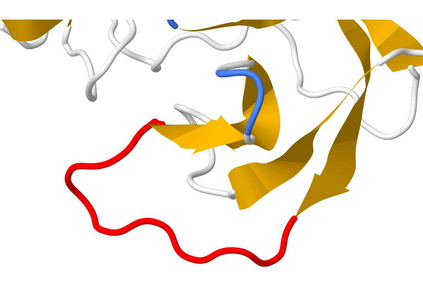
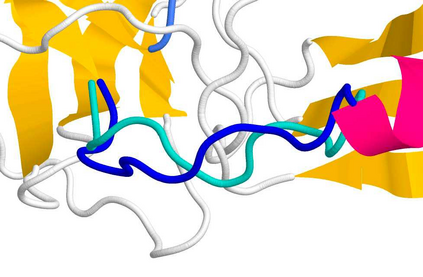
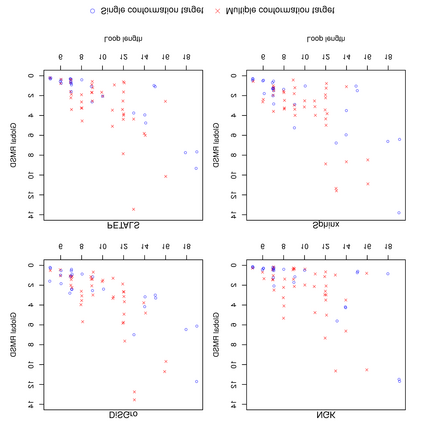
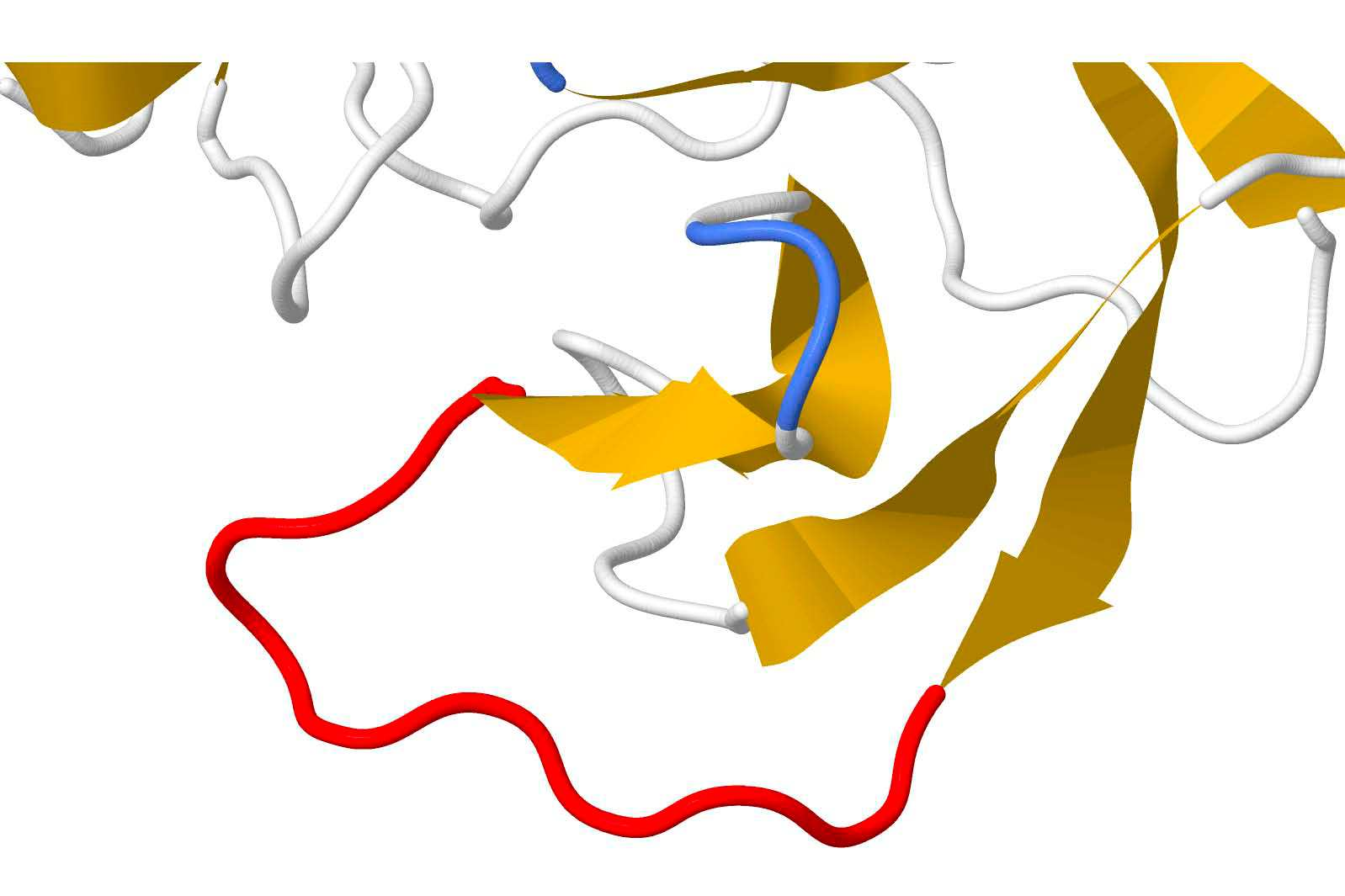
The SARS-CoV-2 spike (S) protein facilitates viral infection, and has been the focus of many structure determination efforts. Its flexible loop regions are known to be involved in protein binding and may adopt multiple conformations. This paper identifies the S protein loops and studies their conformational variability based on the available Protein Data Bank (PDB) structures. While most loops had essentially one stable conformation, 17 of 44 loop regions were observed to be structurally variable with multiple substantively distinct conformations based on a cluster analysis. Loop modeling methods were then applied to the S protein loop targets, and the prediction accuracies discussed in relation to the characteristics of the conformational clusters identified. Loops with multiple conformations were found to be challenging to model based on a single structural template.
翻译:SARS-COV-2(S)峰值蛋白质有助于病毒感染,是许多结构确定努力的重点,已知灵活环状区域涉及蛋白质结合,并可能采用多种异质,本文件根据现有的蛋白质数据库结构确定S蛋白循环,研究蛋白循环的异质性,虽然大多数环状区域基本上有一个稳定的异质,但发现44个环状区域中有17个在结构上可变,在群集分析的基础上有多种实质性不同的相异性。