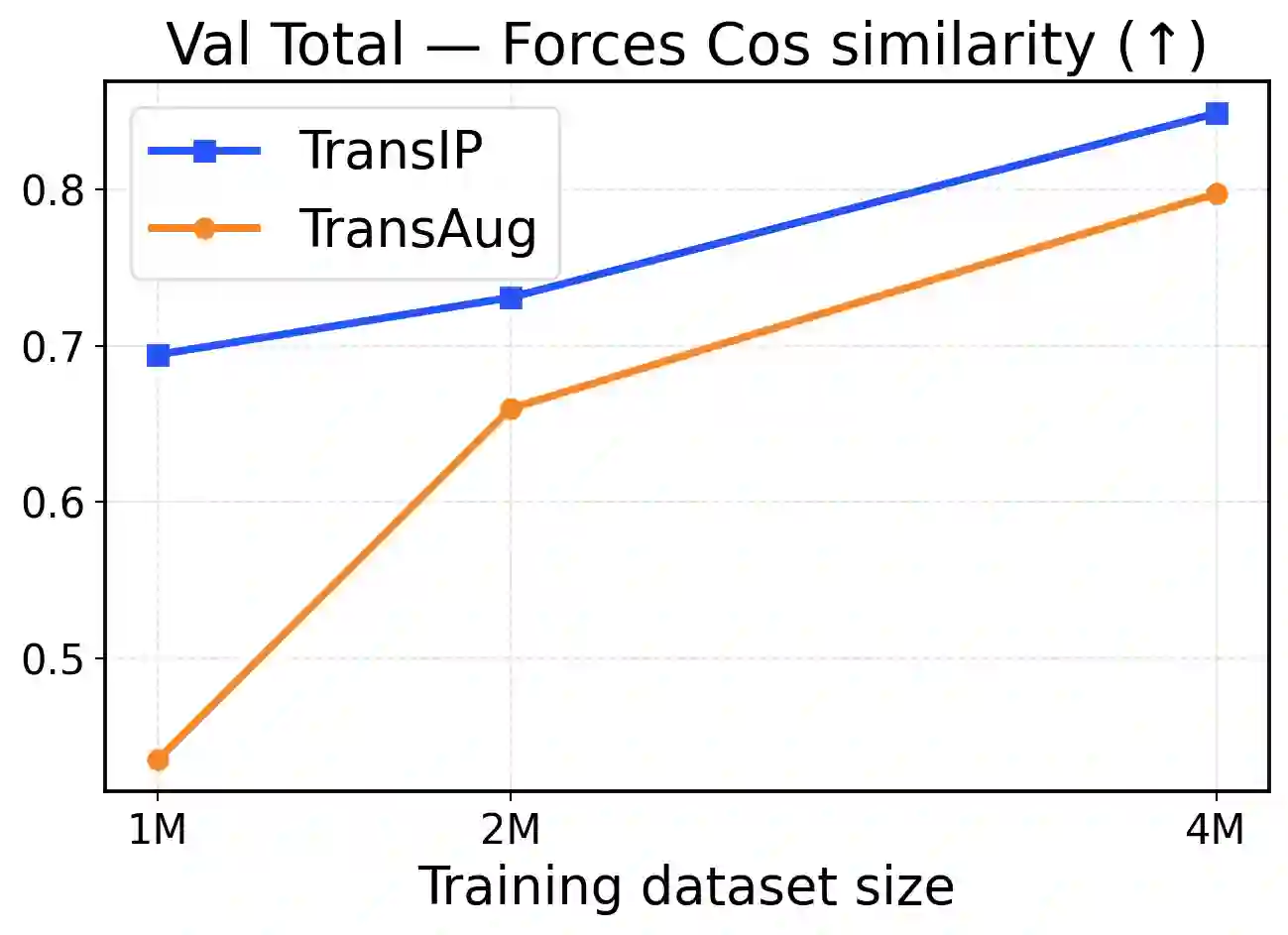
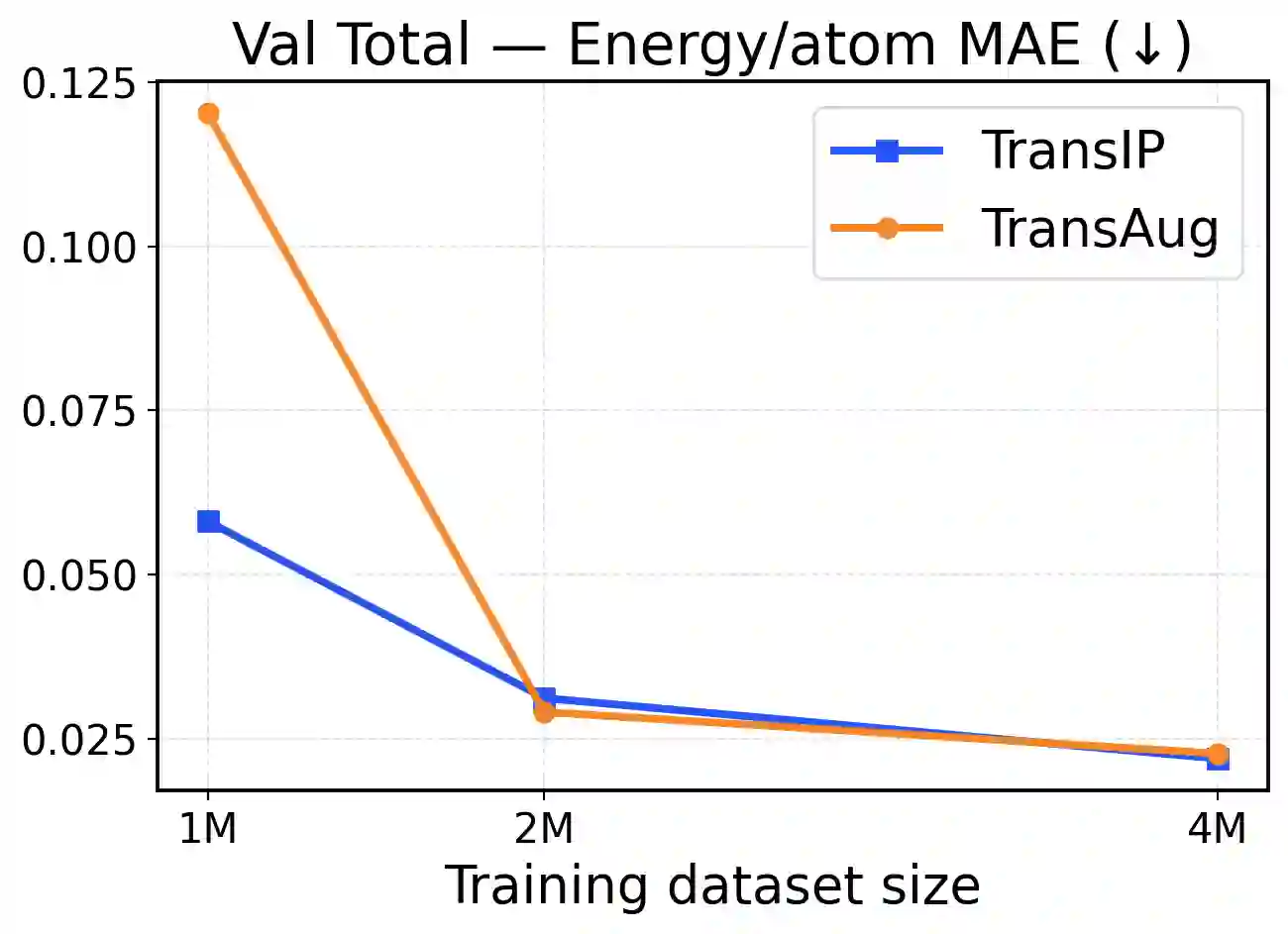
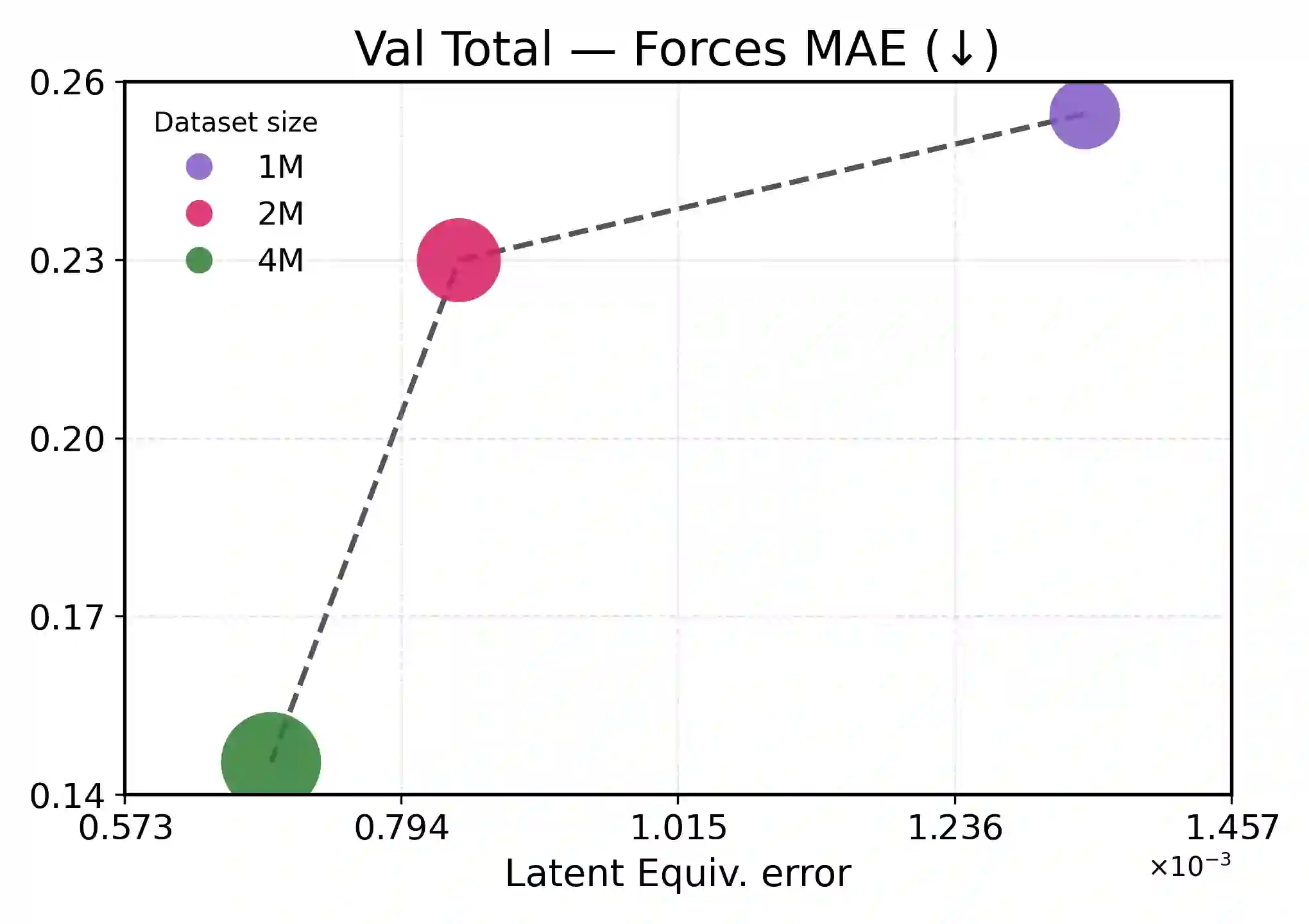
Accurate and scalable machine-learned inter-atomic potentials (MLIPs) are essential for molecular simulations ranging from drug discovery to new material design. Current state-of-the-art models enforce roto-translational symmetries through equivariant neural network architectures, a hard-wired inductive bias that can often lead to reduced flexibility, computational efficiency, and scalability. In this work, we introduce TransIP: Transformer-based Inter-Atomic Potentials, a novel training paradigm for interatomic potentials achieving symmetry compliance without explicit architectural constraints. Our approach guides a generic non-equivariant Transformer-based model to learn SO(3)-equivariance by optimizing its representations in the embedding space. Trained on the recent Open Molecules (OMol25) collection, a large and diverse molecular dataset built specifically for MLIPs and covering different types of molecules (including small organics, biomolecular fragments, and electrolyte-like species), TransIP effectively learns symmetry in its latent space, providing low equivariance error. Further, compared to a data augmentation baseline, TransIP achieves 40% to 60% improvement in performance across varying OMol25 dataset sizes. More broadly, our work shows that learned equivariance can be a powerful and efficient alternative to augmentation-based MLIP models.
翻译:准确且可扩展的机器学习原子间势能(MLIPs)对于从药物发现到新材料设计的分子模拟至关重要。当前最先进的模型通过等变神经网络架构强制实施旋转平移对称性,这种硬编码的归纳偏置常常导致灵活性、计算效率和可扩展性降低。在本研究中,我们提出TransIP:基于Transformer的原子间势能,这是一种实现对称性遵从而无需显式架构约束的新型原子间势能训练范式。我们的方法通过优化嵌入空间中的表示,引导通用的非等变Transformer模型学习SO(3)等变性。基于近期专为MLIP构建的大型多样化分子数据集Open Molecules(OMol25)进行训练(该数据集涵盖多种分子类型,包括小分子有机物、生物分子片段和电解质类物质),TransIP在其潜在空间中有效学习对称性,实现了较低的等变误差。此外,与数据增强基线相比,TransIP在不同规模的OMol25数据集上实现了40%至60%的性能提升。更广泛而言,我们的研究表明,学习型等变性可以成为基于增强的MLIP模型强大而高效的替代方案。