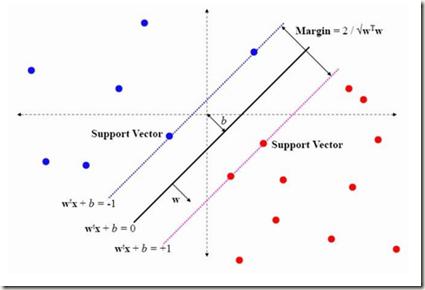
The innovation of next-generation sequencing (NGS) techniques has significantly reduced the price of genome sequencing, lowering barriers to future medical research; it is now feasible to apply genome sequencing to studies where it would have previously been cost-inefficient. Identifying damaging or pathogenic mutations in vast amounts of complex, high-dimensional genome sequencing data may be of particular interest to researchers. Thus, this paper's aims were to train machine learning models on the attributes of a genetic mutation to predict LoFtool scores (which measure a gene's intolerance to loss-of-function mutations). These attributes included, but were not limited to, the position of a mutation on a chromosome, changes in amino acids, and changes in codons caused by the mutation. Models were built using the univariate feature selection technique f-regression combined with K-nearest neighbors (KNN), Support Vector Machine (SVM), Random Sample Consensus (RANSAC), Decision Trees, Random Forest, and Extreme Gradient Boosting (XGBoost). These models were evaluated using five-fold cross-validated averages of r-squared, mean squared error, root mean squared error, mean absolute error, and explained variance. The findings of this study include the training of multiple models with testing set r-squared values of 0.97.
翻译:暂无翻译