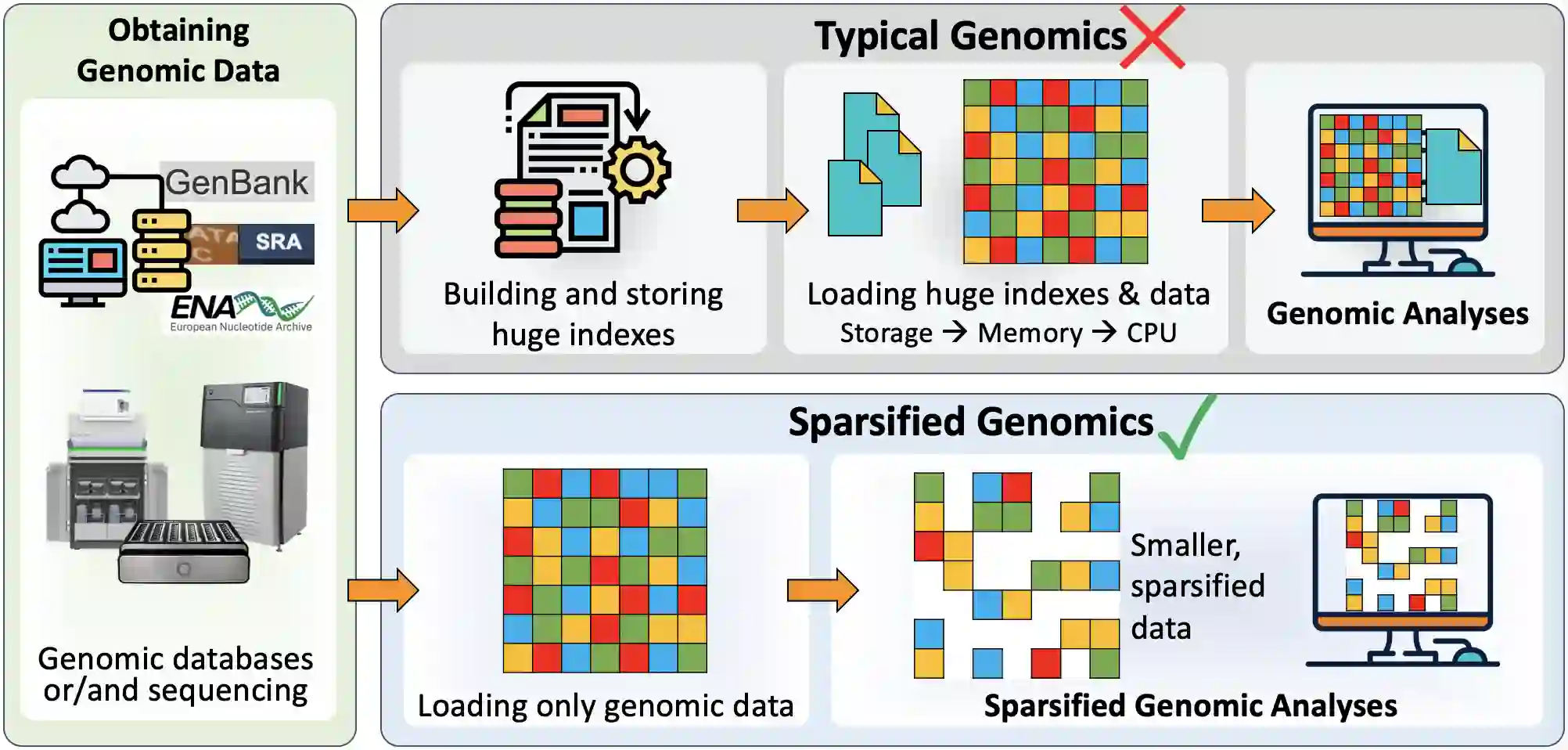
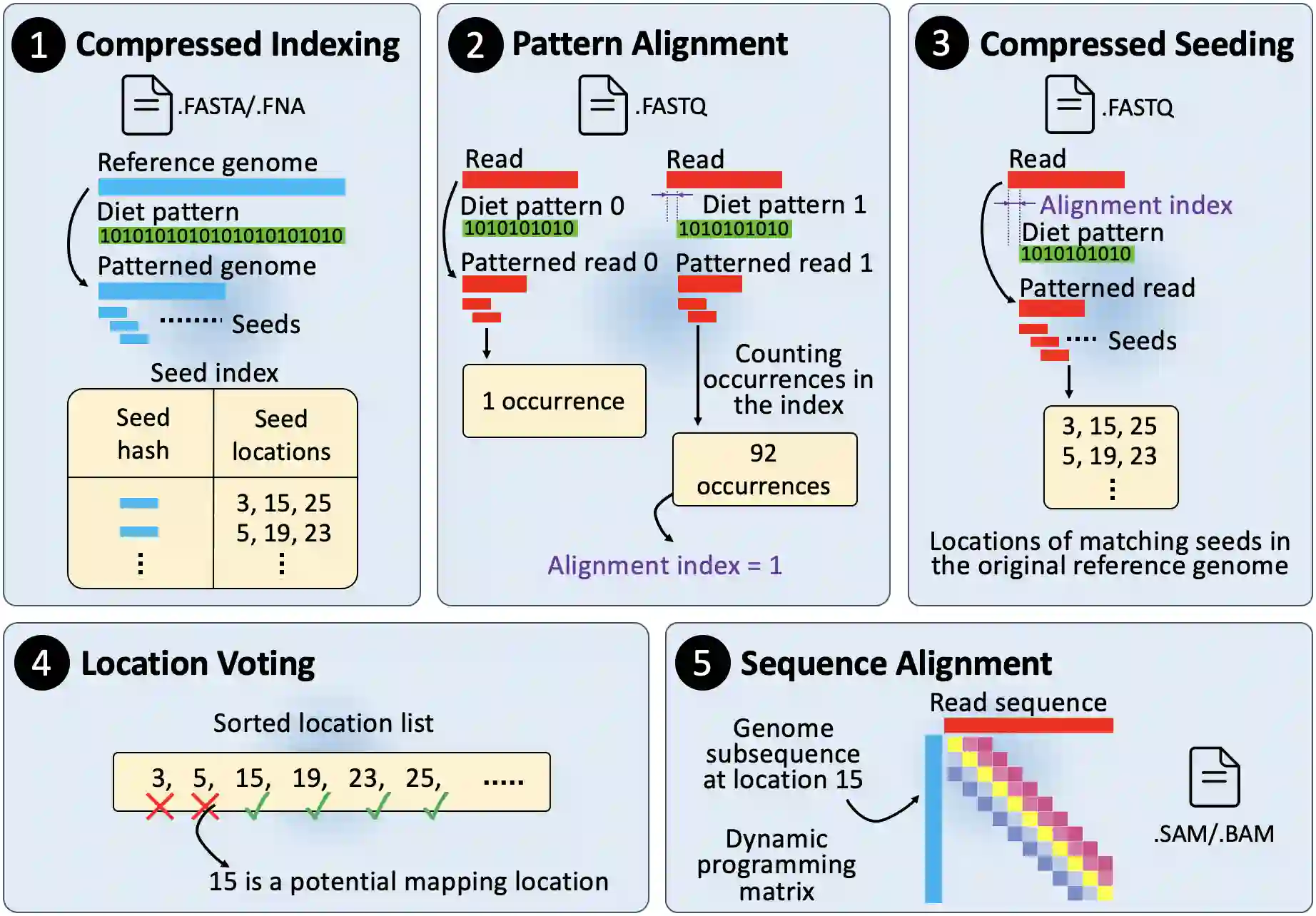
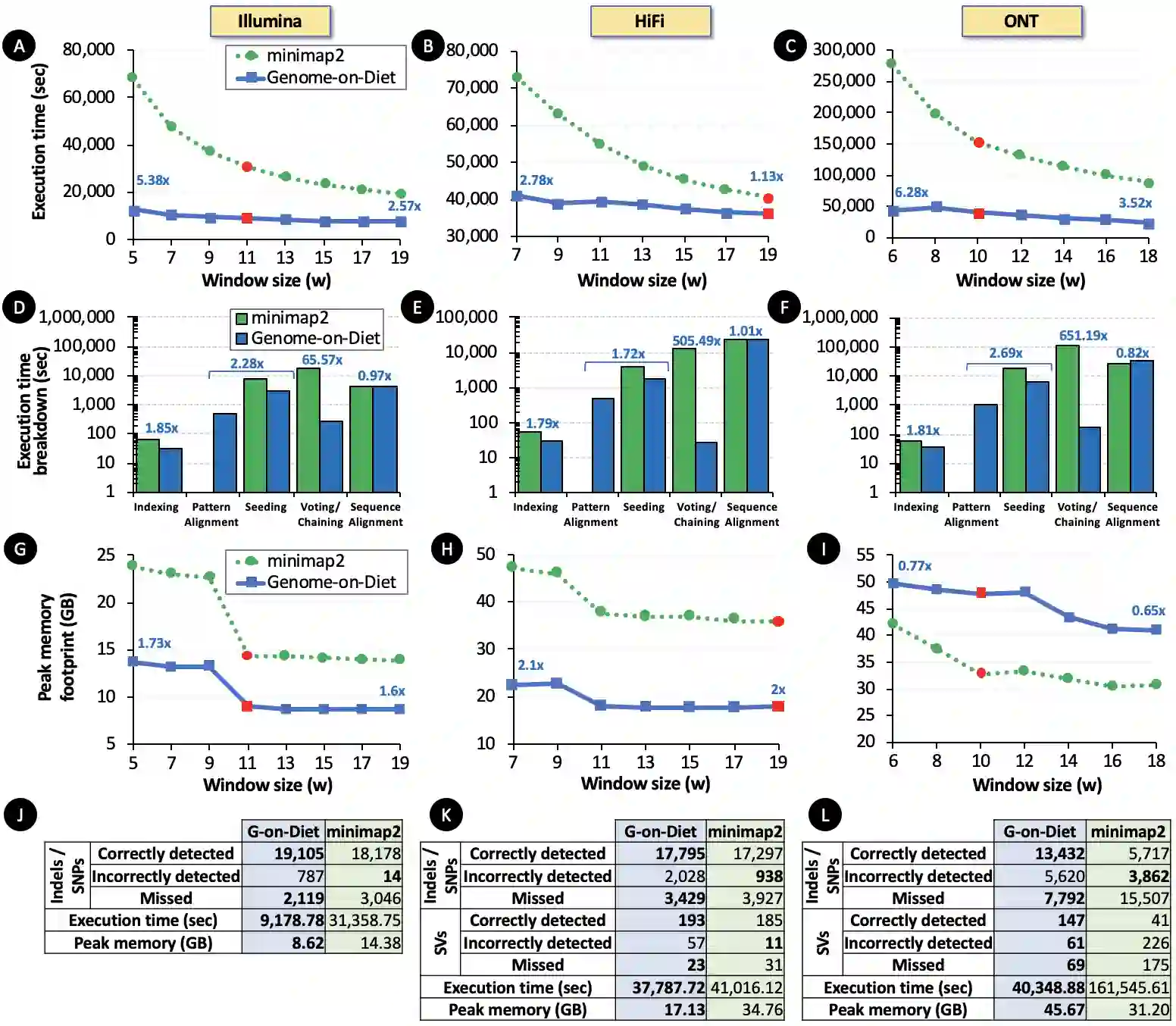
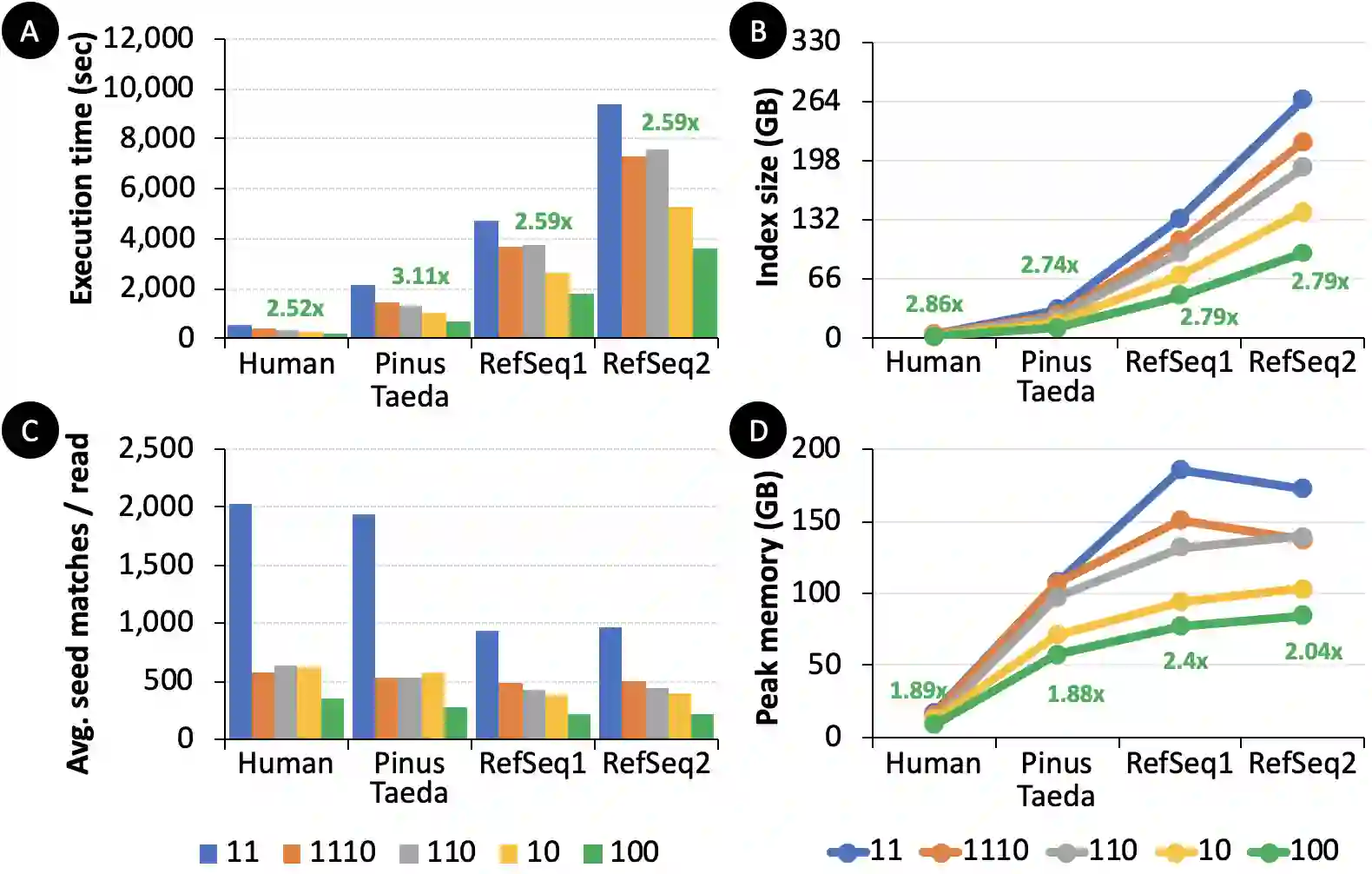
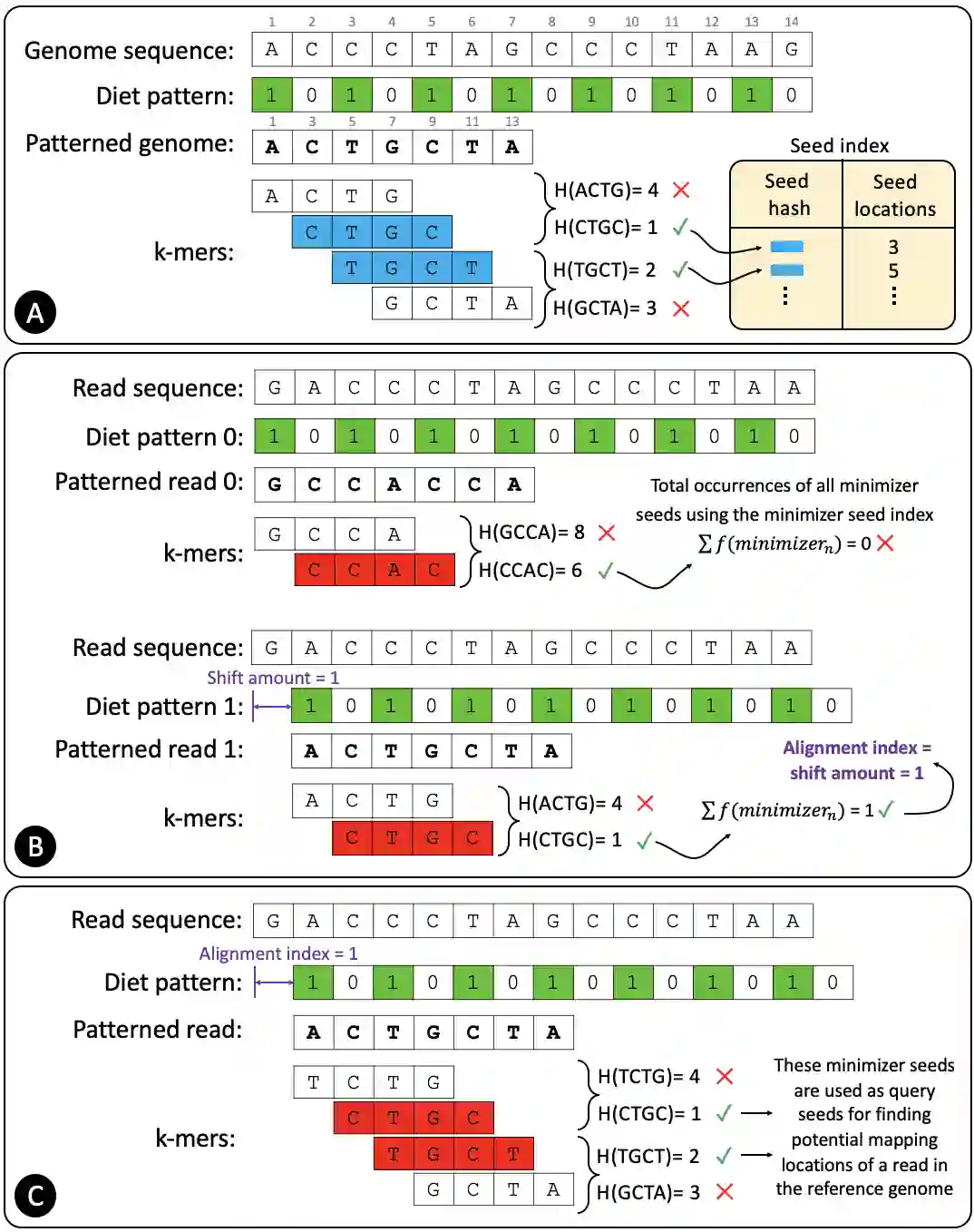
Searching for similar genomic sequences is an essential and fundamental step in biomedical research and an overwhelming majority of genomic analyses. State-of-the-art computational methods performing such comparisons fail to cope with the exponential growth of genomic sequencing data. We introduce the concept of sparsified genomics where we systematically exclude a large number of bases from genomic sequences and enable much faster and more memory-efficient processing of the sparsified, shorter genomic sequences, while providing similar or even higher accuracy compared to processing non-sparsified sequences. Sparsified genomics provides significant benefits to many genomic analyses and has broad applicability. We show that sparsifying genomic sequences greatly accelerates the state-of-the-art read mapper (minimap2) by 2.57-5.38x, 1.13-2.78x, and 3.52-6.28x using real Illumina, HiFi, and ONT reads, respectively, while providing up to 2.1x smaller memory footprint, 2x smaller index size, and more truly detected small and structural variations compared to minimap2. Sparsifying genomic sequences makes containment search through very large genomes and very large databases 72.7-75.88x faster and 723.3x more storage-efficient than searching through non-sparsified genomic sequences (with CMash and KMC3). Sparsifying genomic sequences enables robust microbiome discovery by providing 54.15-61.88x faster and 720x more storage-efficient taxonomic profiling of metagenomic samples over the state-of-art tool (Metalign). We design and open-source a framework called Genome-on-Diet as an example tool for sparsified genomics, which can be freely downloaded from https://github.com/CMU-SAFARI/Genome-on-Diet.
翻译:在生物医学研究和绝大多数基因组分析中,寻找类似的基因组序列是一个基本和根本的步骤。进行这种比较的最先进的计算方法无法应付基因组序列数据的指数增长。我们引入了环球基因组学概念,我们系统地将大量基数从基因组序列中排除出来,并能够更快和更快地进行存储效率更高的处理,同时提供与处理非基因组序列相比的类似或甚至更高的准确性。经过精密的基因组学为许多基因组分析提供了重大好处,并且具有广泛适用性。我们表明,通过SMA355-5.38x、1.13-2.78x和3.52-6.28x, 利用真正的光学、HiFi和ONT等序列进行快速和记忆效率更高的处理,同时提供比MINGMIS488更小的存储和结构变异异性数据。 通过SMA353的大规模搜索数据序列设计,SMADRM-M-S-MIS-MS-M-M-Smex, 更快速的SMI-MI-MI-S-MIS-MIS-MIS-MIS-I-I-S-MIS-MIOL-MIS-S-M-S-S-S-SIMOL-S-S-S-SIMOL-IL-IL-S-S-S-S-S-S-SM-S-S-S-S-S-S-S-M-S-S-S-S-S-S-S-SM-M-SM-M-SM-SM-M-SD-M-SD-SD-SD-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-SM-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-S-