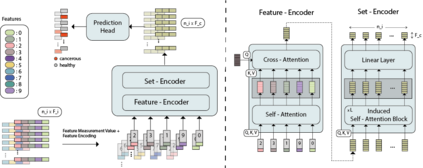
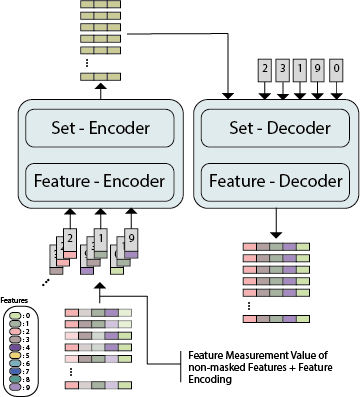
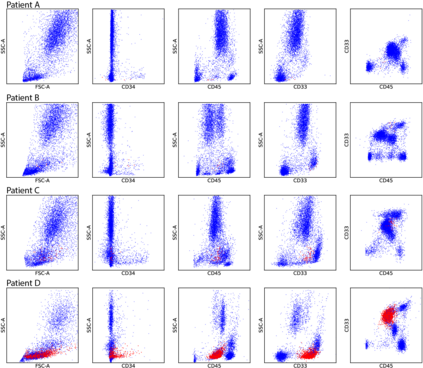
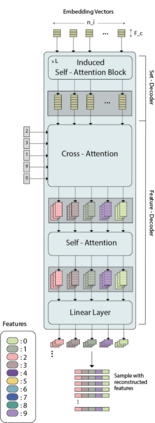
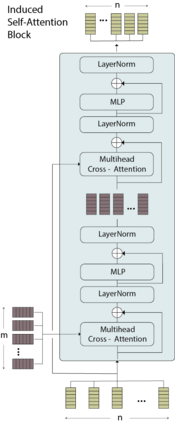
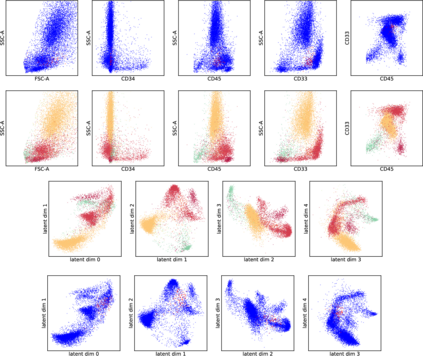
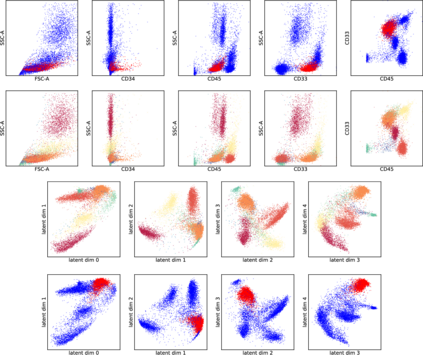
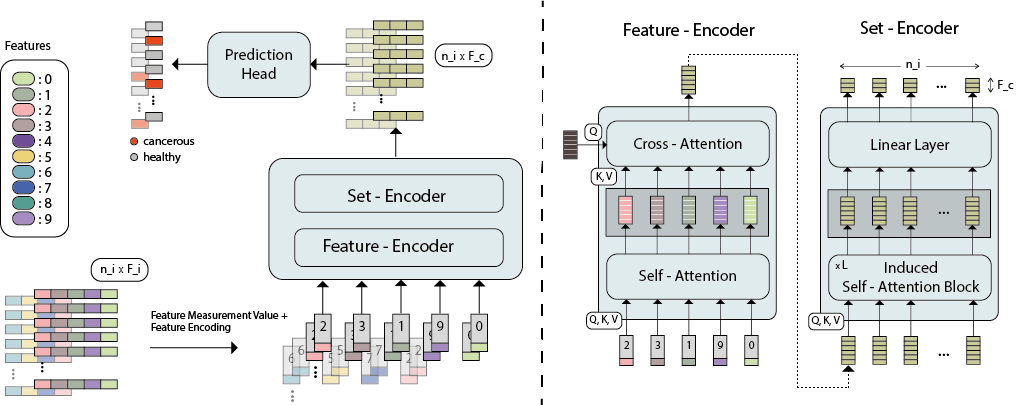
While model architectures and training strategies have become more generic and flexible with respect to different data modalities over the past years, a persistent limitation lies in the assumption of fixed quantities and arrangements of input features. This limitation becomes particularly relevant in scenarios where the attributes captured during data acquisition vary across different samples. In this work, we aim at effectively leveraging data with varying features, without the need to constrain the input space to the intersection of potential feature sets or to expand it to their union. We propose a novel architecture that can directly process data without the necessity of aligned feature modalities by learning a general embedding space that captures the relationship between features across data samples with varying sets of features. This is achieved via a set-transformer architecture augmented by feature-encoder layers, thereby enabling the learning of a shared latent feature space from data originating from heterogeneous feature spaces. The advantages of the model are demonstrated for automatic cancer cell detection in acute myeloid leukemia in flow cytometry data, where the features measured during acquisition often vary between samples. Our proposed architecture's capacity to operate seamlessly across incongruent feature spaces is particularly relevant in this context, where data scarcity arises from the low prevalence of the disease. The code is available for research purposes at https://github.com/lisaweijler/FATE.
翻译:暂无翻译