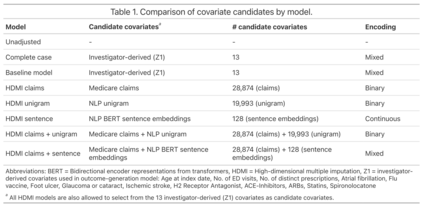
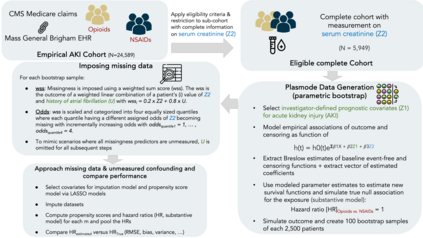
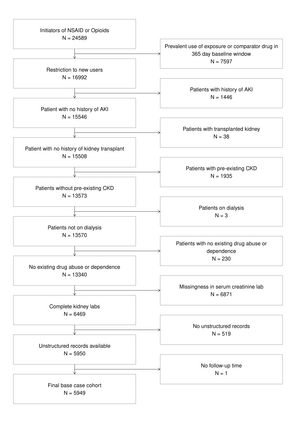
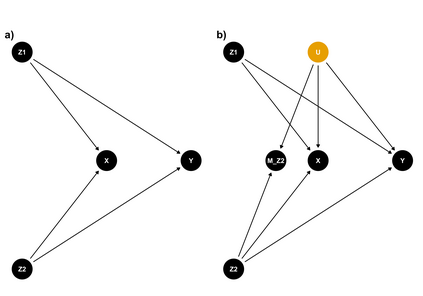
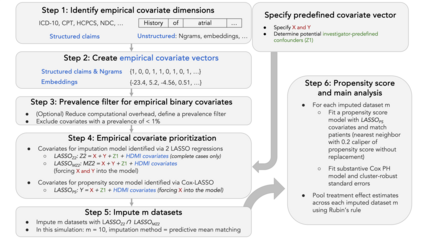
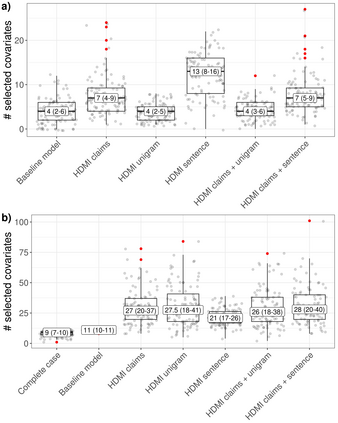
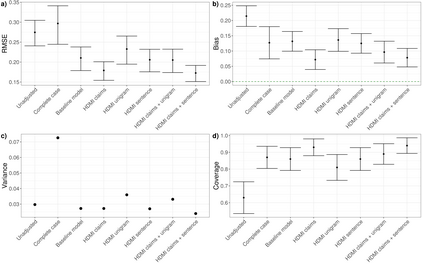
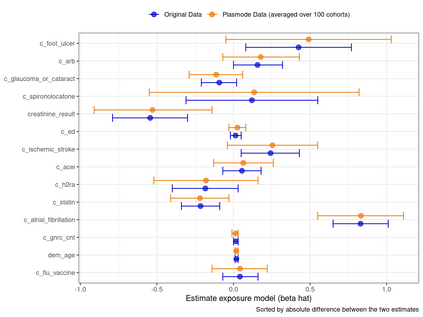
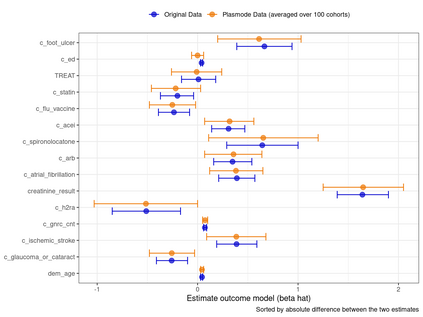
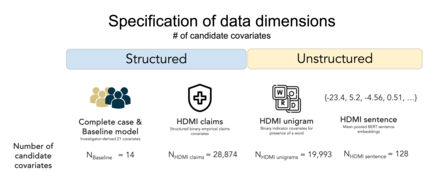
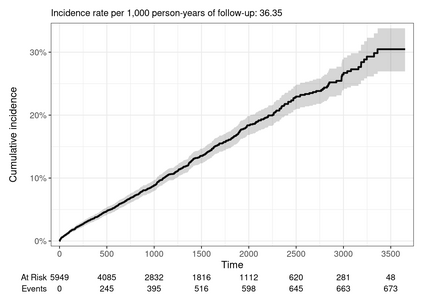
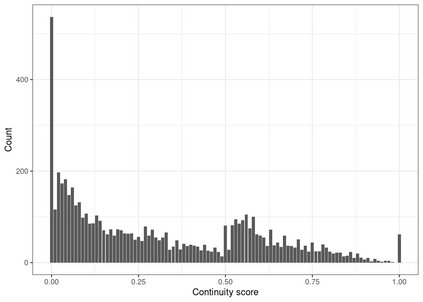
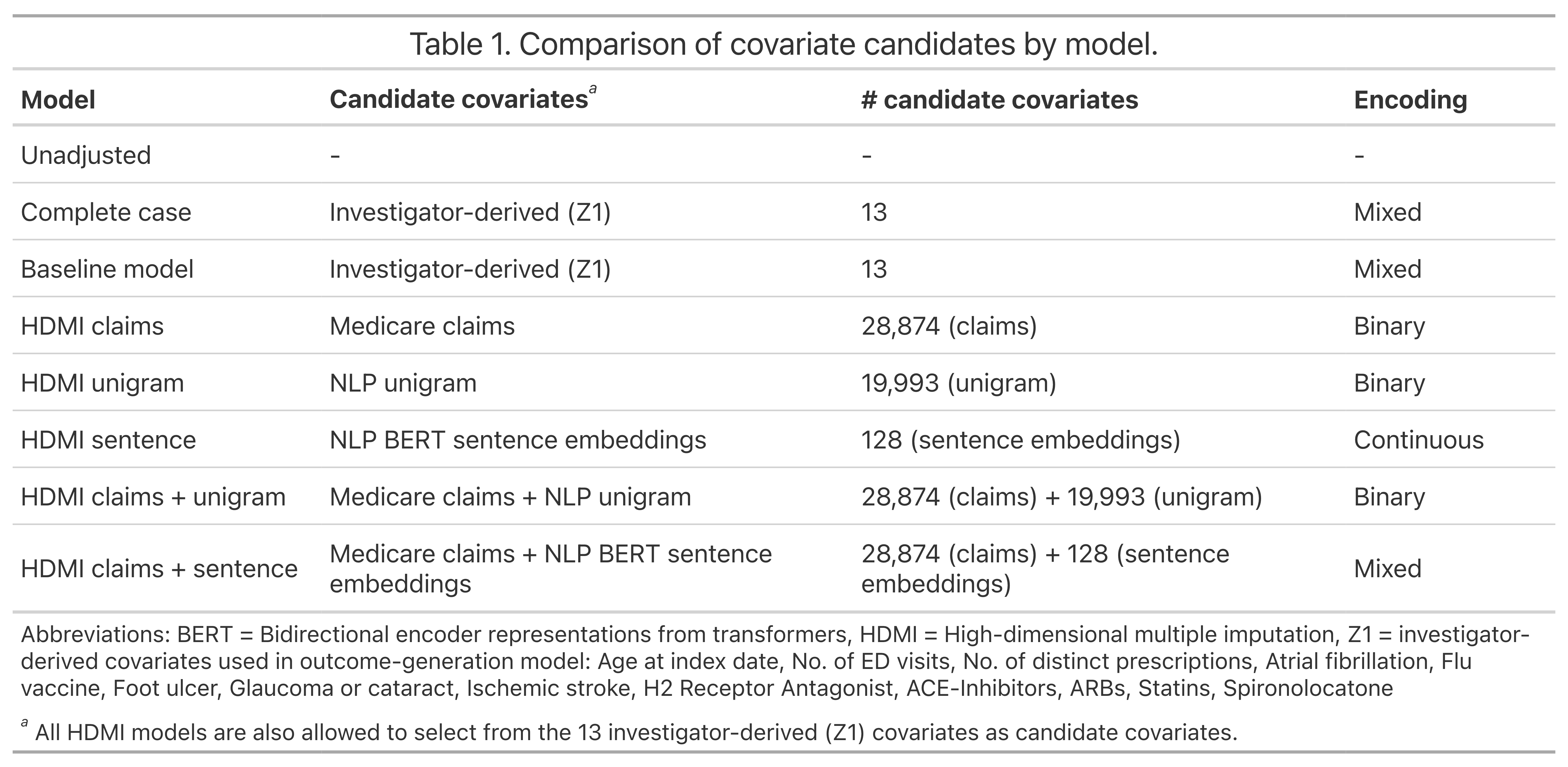
Multiple imputation (MI) models can be improved by including auxiliary covariates (AC), but their performance in high-dimensional data is not well understood. We aimed to develop and compare high-dimensional MI (HDMI) approaches using structured and natural language processing (NLP)-derived AC in studies with partially observed confounders. We conducted a plasmode simulation study using data from opioid vs. non-steroidal anti-inflammatory drug (NSAID) initiators (X) with observed serum creatinine labs (Z2) and time-to-acute kidney injury as outcome. We simulated 100 cohorts with a null treatment effect, including X, Z2, atrial fibrillation (U), and 13 other investigator-derived confounders (Z1) in the outcome generation. We then imposed missingness (MZ2) on 50% of Z2 measurements as a function of Z2 and U and created different HDMI candidate AC using structured and NLP-derived features. We mimicked scenarios where U was unobserved by omitting it from all AC candidate sets. Using LASSO, we data-adaptively selected HDMI covariates associated with Z2 and MZ2 for MI, and with U to include in propensity score models. The treatment effect was estimated following propensity score matching in MI datasets and we benchmarked HDMI approaches against a baseline imputation and complete case analysis with Z1 only. HDMI using claims data showed the lowest bias (0.072). Combining claims and sentence embeddings led to an improvement in the efficiency displaying the lowest root-mean-squared-error (0.173) and coverage (94%). NLP-derived AC alone did not perform better than baseline MI. HDMI approaches may decrease bias in studies with partially observed confounders where missingness depends on unobserved factors.
翻译:暂无翻译