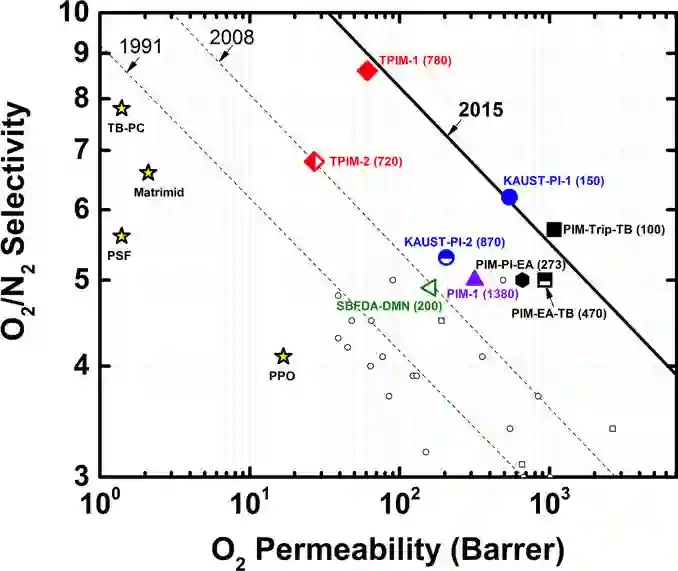
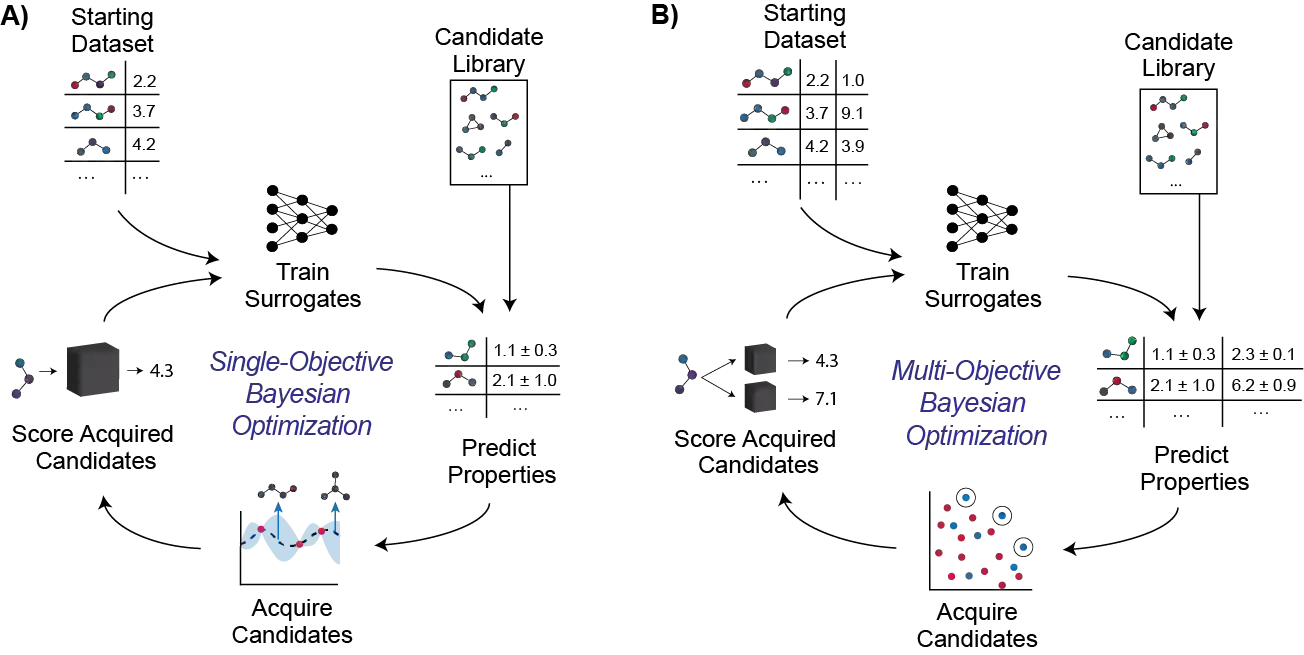
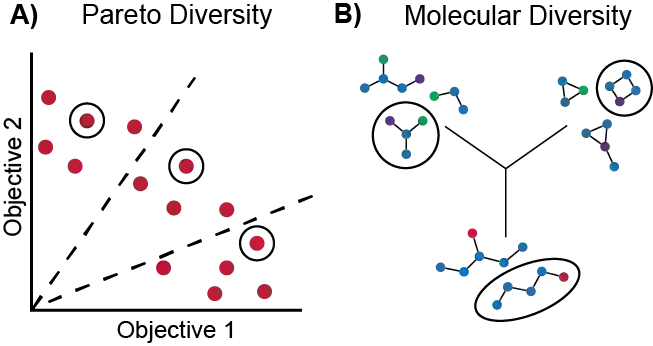
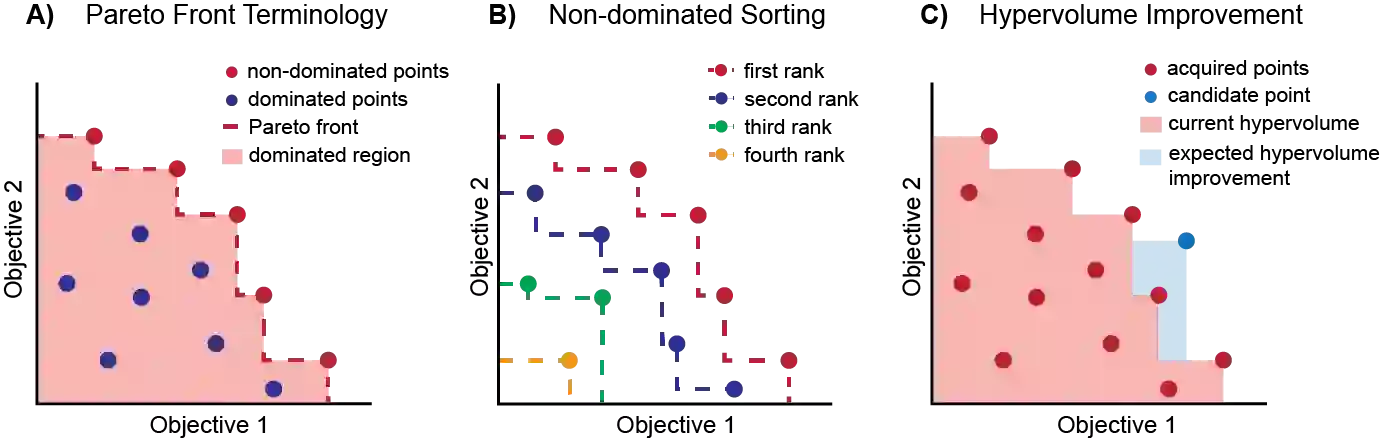Molecular discovery is a multi-objective optimization problem that requires identifying a molecule or set of molecules that balance multiple, often competing, properties. Multi-objective molecular design is commonly addressed by combining properties of interest into a single objective function using scalarization, which imposes assumptions about relative importance and uncovers little about the trade-offs between objectives. In contrast to scalarization, Pareto optimization does not require knowledge of relative importance and reveals the trade-offs between objectives. However, it introduces additional considerations in algorithm design. In this review, we describe pool-based and de novo generative approaches to multi-objective molecular discovery with a focus on Pareto optimization algorithms. We show how pool-based molecular discovery is a relatively direct extension of multi-objective Bayesian optimization and how the plethora of different generative models extend from single-objective to multi-objective optimization in similar ways using non-dominated sorting in the reward function (reinforcement learning) or to select molecules for retraining (distribution learning) or propagation (genetic algorithms). Finally, we discuss some remaining challenges and opportunities in the field, emphasizing the opportunity to adopt Bayesian optimization techniques into multi-objective de novo design.
翻译:分子发现是一个多客观的优化问题,它要求确定一个分子或一组分子,平衡多种、往往是相互竞争的特性。多客观的分子设计通常通过使用缩放法将感兴趣的特性结合成单一的客观功能来解决,这种剖析法对相对重要性的假设没有多少发现,对目标之间的取舍也很少发现。与缩放法相比,Pareto优化并不要求了解相对重要性,而是揭示了目标之间的取舍。然而,它提出了算法设计中的额外考虑。在本次审查中,我们描述了以Pareto优化算法为焦点的多目标分子发现以集合为基础和无异基因化方法进行。我们展示了基于集合分子的发现是如何将多目标巴耶斯优化法相对直接延伸的,以及不同基因化模型从单一目标到多目标优化的方式如何以类似的方式扩展,使用非主排序功能(强制学习)或选择分子用于再培训(分配学习)或传播(遗传算法)的。最后,我们讨论了该领域中仍然存在的一些挑战和机遇,强调采用巴伊斯最佳设计技术的机会。